4 Reads Per Cluster! Simultaneously Paired End-Sequencing!
Executive Summary: Illumina have a method which could double the throughput of their instruments. But it would most likely result in slightly lower accuracy… at least as implemented on current sequencers.
gdub42 has been posting some interesting Illumina patents over on the Discord. One of these is an interesting approach describing simultaneous readout of both the first and second read at the same time.
This would nearly half sequencing run times, giving Illumina another edge over the new crop of sequencers that are appearing.
Background
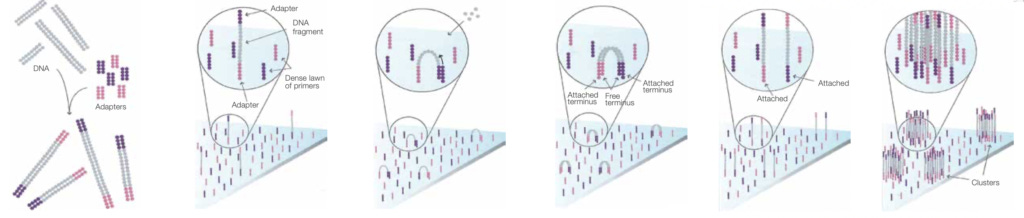
Illumina flowcells are patterned with two different primers (P5 and P7) these are attached adapters on your templates used during the bridge amplification process:
During the bridge amplification process we create forward and reverse strands from the original template. So each cluster contains two different types of strands:
Prior to sequencing all those reverse strands are cleaved off and washed out. Leaving you with a cluster containing only the forward strands.
Why do we cleave off those reverse strands? Well. I imagine they would probably transiently hybridize and generally be a pain to have around. In any case we get rid of them.
Then we prime for sequencing and read out the forward strand. We could just stop there… and many users do.
But we still have those other primers on the flowcell, so it’s possible to extract more information out of our templates.
So… we regenerate the reverse strands, perform some more amplification cycles and this time cleave off the forward strands. We can now prime these reverse strands and sequence in the opposite direction.
Double the reads for your money!
What If We Did Both At The Same Time?
In the discord we were discussing the possibility of doing both the forward and reverse read at the same time.
If you had some way of only using one set of colors on the first read and another set of colors on the second read you could sequence them both at the same time!
The problem is we don’t have that. Nucleotides will just incorporate wherever they want. We can’t say read 1 will incorporate only red and green nucleotides, and read 2 blue and yellow.
Illumina have therefore been looking at a different solution to the problem.