Chemistry X and Long Reads
Of Illumina’s recent announcements, Chemistry X seemed the most interesting to me. Not really because this is a significant leap forward, but because of what it says about the fundamental limitations of cluster based sequencing-by-synthesis…
If recent Illumina patents are anything to go by, prephasing (multiple-incorporation) and phasing (non-incorporation) are both down at the <0.1% level. But as previously discussed, bleaching seems to very low. With dye intensities staying high until the end of the run:
Ultimately, this suggests that phasing therefore is now the limiting factor for read length. Over on twitter, someone suggested that as phasing is a (somewhat) deterministic process, can we not completely correct for it?
The issue is however, that even at these modest (0.1%) levels, strands get so out of phase, that information is lost and you can no longer accurately determine the original sequence.
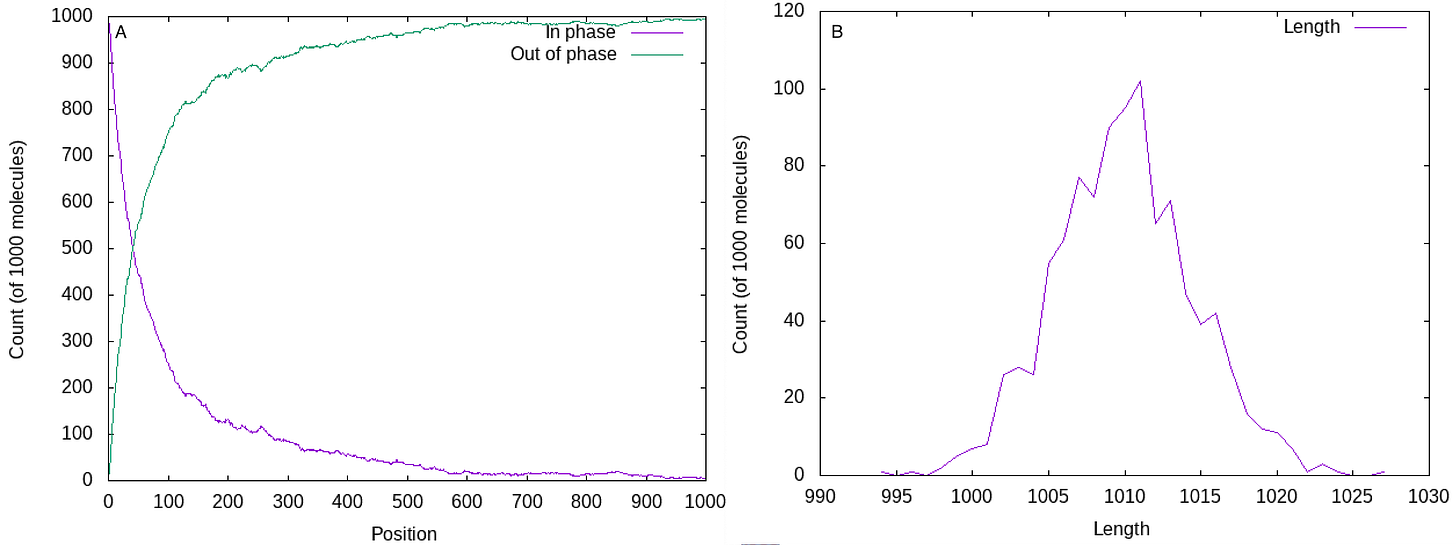
I threw together a quick simulation of a cluster containing 1000 strands. The figure below shows what happens at cycle 1000 with 1% phasing/prephasing is applied. By cycle 1000, 92.2% of strands are “out of phase”:
If we look at how far each of the strands has advanced (part B above), we can see the average length is still around 1000. But the FWHM is about 10. Our signal is spread across >10 positions. And this is actually the ideal case where phasing and prephasing are balanced and phasing and prephasing tend to cancel each other out.
If we simulate unbalanced prephasing/phasing, a much smaller fraction of strands remain ‘in phase”:
Pushing this down to a 0.1% we’re still ~30% out of phase at cycle 1000:
So, while the information is there, it’s spread across multiple cycles, and when we’re down to clusters of a few thousand strands. Individual contributions will be undetectable. This therefore limits read length, and if the Chemistry X announcement is anything to go by only provides useful accuracy out to ~600bp.
Behind the paywall below, I’m going to discuss an approach which might allow long reads to be extracted from these clusters. For any non-subscribers, feel free to reach out and I’ll provide a copy.