Element Biosciences - Further Thoughts
A few things came up after my last post on Element. Firstly, over on twitter it was noted that Element have said they use a “multivalent binding” approach and Simon Barnett pointed me toward this patent, which seems like a likely candidate.
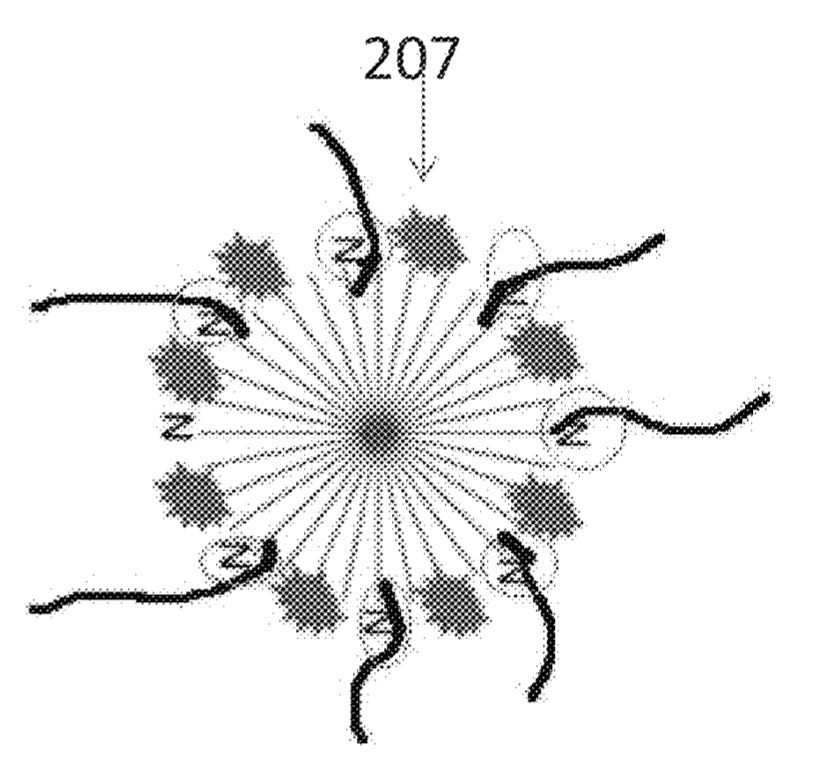
The patent proposes the use of particle-nucleotide conjugates. To my non-chemist eye, this is a bunch of nucleotides stuck together with fluorophores:
Much like the nucleotide-polymerase conjugate approach, this can be used in a two step process. As previously described, the conjugates may be contacted and bind, but not incorporate. Then after imaging, removed and regular reversible terminators used to advance the strand to the next position:
I assume that by using this big ball of nucleotides binding to multiple strands in a nanoball, you potentially get stronger binding. Perhaps you also have more flexibility in terms of your nucleotide-to-fluorophore ratio. This could potentially enable shorter exposure times…
However I’d imagine this approach potentially introduces some new issues. The number of fluorophores/bond conjugates should be relatively tightly controlled. If the number of active fluorophores/sites is highly variable this could introduce noise making it harder to estimate and correct for phasing. Maybe their approach has inherently less phasing anyway…
Omniome and Element
As mentioned in my previous post this two step approach still seem quite similar to that proposed by Omniome. While I’m no patent lawyer, many of the claims in this patent:
“The method of claim 1, wherein the polymerase molecules are catalytically inactive.”
“The method of claim 1, wherein the contacting is done in the presence of strontium ions, magnesium ions, calcium ions, or any combination thereof.”
“The method of claim 1, wherein the polymerase molecules have been rendered catalytically inactive by the absence of a necessary ion or cofactor”
Seem somewhat similar to those in some of Omniome’s:
“The method of claim 13, where in the stabilizing agent is a non-catalytic metal ion.”
“The method of claim 14, where in the non-catalytic metal ion is strontium, tin, or nickel.”
I suppose it all depends on the details, and exactly what mechanism is being used to deactivate the polymerase. Element do also mention using engineered Polymerases that bind but don’t incorporate, could be worth hunting through their patents for sequence listings (though I didn’t see any while scanning through).
Imaging
Finally, I had a quick poke around in some of Element’s imaging related patents. Overall the approach seems pretty standard, and suggests that they are imaging both sides of the flowcell. As I understand it, Illumina also do this on at least some of their instruments:
Re-reading Keith’s post, I also noticed he specifically asked if they were using patterned flowcells and it seems they are not. Figures from Element patents also all seem to show typical 4 channel imaging systems (rather than Illumina-style 2-color):
So, overall things seem somewhat similar to Singular (and early Illumina instruments) on the optical side. It’s therefore not clear to me why they have a throughput advantage over Singular, on a per flowcell basis.
I’m sure as more is revealed about these platforms all will become clear!