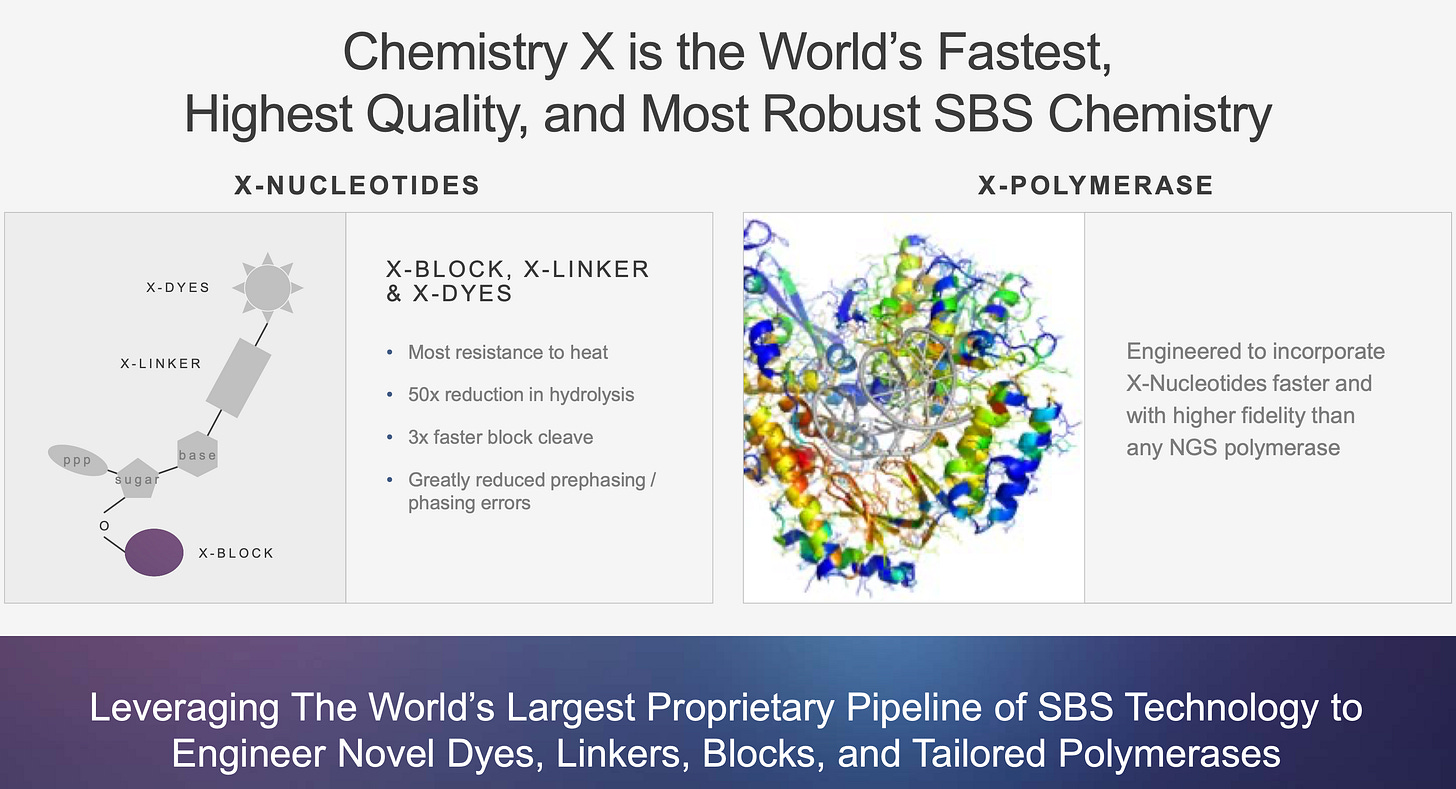
Illumina - Chemistry X
At JPM Illumina presented “Chemistry X” which they suggested will result in 2x longer reads. Kevin, over at OmicsOmics has some interesting thoughts on this, and its utility.
Unlike Infinity, these are real actual longer reads, rather than synthetic ones.
2x longer reads would suggest 500 to 600bp single end reads (Miseq 300bp, Novaseq 250bp).
Before I speculate on Chemistry X, let’s review the issues that prevent Illumina’s current chemistry from extending beyond 300bp. There are two main issues here, phasing and bleaching.
Phasing
Phasing is better described elsewhere, but essentially is the tendency for strands to get out of sync. Either a strand fails to incorporate a nucleotide, or it incorporates multiple nucleotides. This results in a smeared signal where you see signal bleeding through to adjacent cycles. The Illumina primary data analysis pipeline contains corrections for phasing artifacts. However as phasing builds up the performance of these corrections goes down, limiting overall read length.
Better incorporation efficiency, and improved blocking efficiency would decrease phasing errors and help increase read length.
Bleaching
Here I’m using bleaching to refer to “loss of signal”. This can be through a number of mechanisms, but most commonly this would be due to photo-damage. This is where illumination causes damage, resulting in a loss of signal.
For example, photo-damage could occur in the strand under synthesis, resulting no further signal from that strand and an overall decrease in signal. Bleaching has a knock on effect on phasing corrections, as if you have less signal, you can’t estimate and correct for phasing as well.
If we look at the intensity (signal) drop across cycle on Illumina runs you’ll generally see a significant drop in intensity across cycle, for example in this plot from an older Miseq run:
Both phasing and bleaching get “reset” with the second read, but limit your single read, read length.
Chemistry X
Illumina said Chemistry X is composed of new nucleotides and a new polymerase. Illumina are always patenting new polymerases so I tend to discount this. However new nucleotides are interesting, particularly as the foundational Solexa nucleotide IP is expiring this year. New nucleotide IP would help give Illumina an edge over other players looking to build on this expired IP.
After this announcement a reader kindly pointed me toward this recent Illumina patent. This may, or may not, be related to Chemistry X. But it seems worth quickly reviewing some of the results shown.
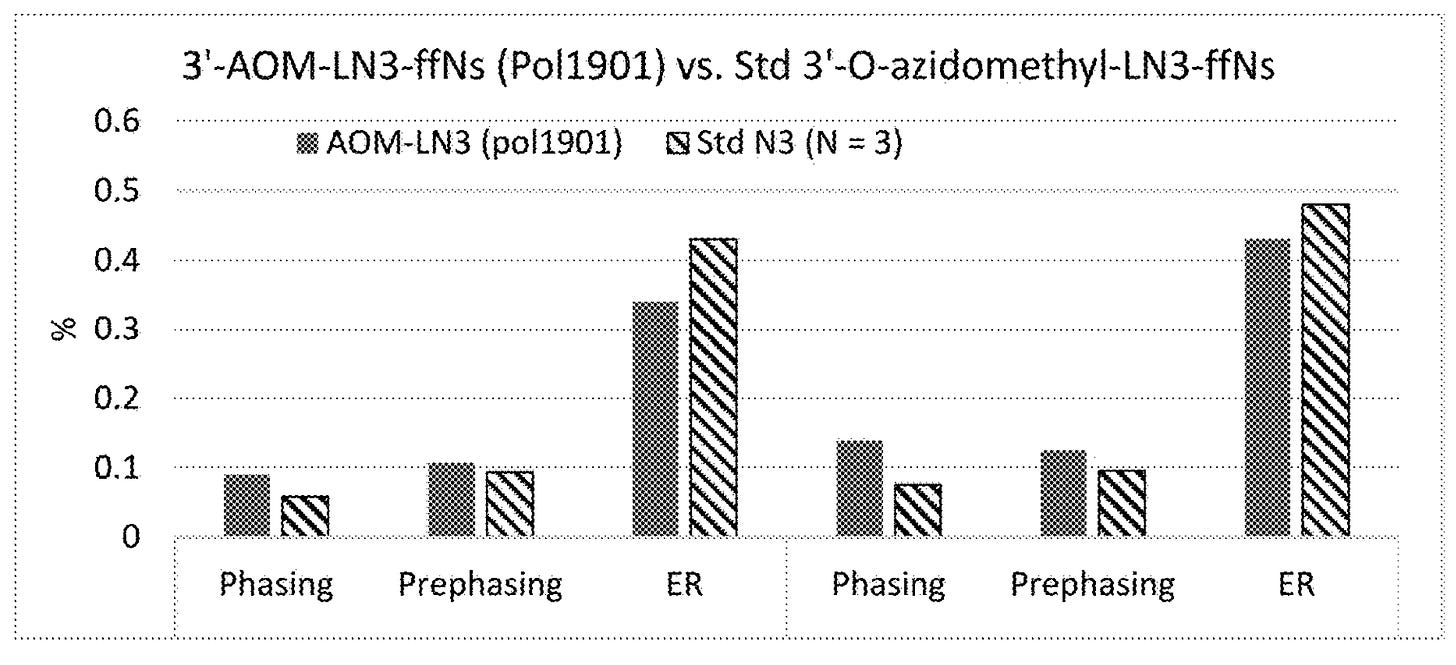
The patent states that “the 3’-OH blocking groups described herein may also achieve low pre-phasing, lower signal decay for improved data quality, which enables longer reads”.
Looking at the graphs, the new (AOM) nucleotides in general show similar phasing and prephasing to those currently used:
However while phasing looks similar or worse, error rates (ER above) are reduced. How can this be? I suspect it’s down to the improved stability these nucleotides are showing. And a plot of intensity across cycle shows essentially “no signal decay”:
As mentioned above, with more signal we can better estimate and correct for phasing artifacts. So it could be that this improved stability helps enable longer read lengths.
But even with prefect stability, if these nucleotides don’t show improved phasing characteristics I suspect this will become the limiting factor.
A hand wavy way of thinking about this might be as follows. Currently we have 50% signal drop, and ~50% of molecules “out of phase” at the end of a read. With the new chemistry we have no signal drop, but the same phasing issues. We can therefore deal with roughly twice as much phasing before our corrections break down. Roughly doubling read length.
This kind of makes sense to me. But certainly suggests that to push beyond 600bp (while maintaining data quality) Illumina will need to further address phasing issues.