Ionic Current Solid State Nanopores
Yesterday I put together some notes on biological ionic current nanopores. Today I’m going to talk about a related approach, solid state ionic current nanopores.
A Background on Solid State Ionic Current Sensing
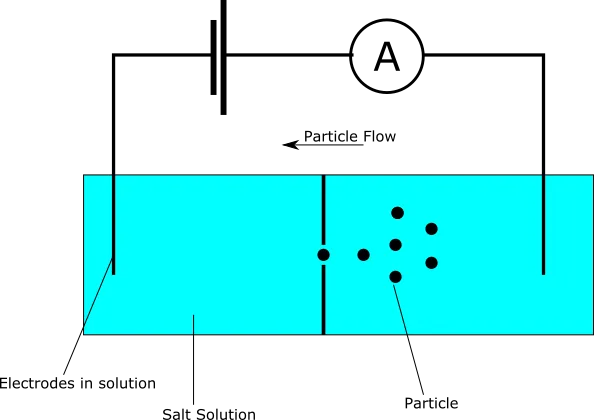
Our experimental setup here is similar to that used for biological pores, we’re just replacing the biological pore with a manufactured one.
As before we have a conductive buffer solution, bias voltage, and current sense amplifier. Particles are detected as they move through a nanoscale hole (nanopore) blocking current flow:
This general principal is really old, and works for particles both large and small. The approach was first used in the 1940s for counting white blood in Coulter counters. Here, whole cells pass through micron scale apertures. The principal even works for much larger particles up to millimeters in size and can be used for measuring particle sizes in paints and for other industrial applications.
In general, the particles used with Coulter counters need to be forced through the “micropores” under pressure driven flow. Some effort needs to be made to keep the flow consistent, and novel pumping methods are often used.
This isn’t the case with DNA. Here the charge on the polymer is large enough that strands will move through the pore under a bias voltage.
Pore Sizes
Clearly, Coulter counter sized pores (microns) will be too large to detect single molecules of DNA. We need a pore that is small enough that the background current doesn’t overwhelm any current blockage, and where the DNA strand is somewhat confined in terms of its conformation and orientation as it passes through the pore.
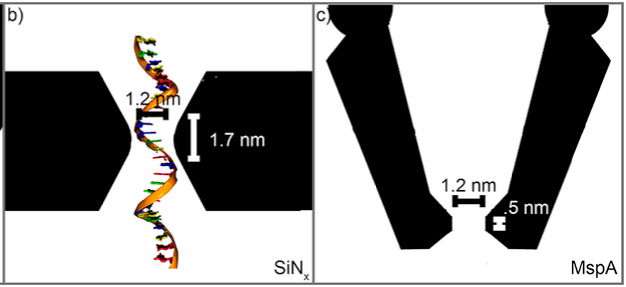
Before we discuss experimental results, we can get some idea of the feature sizes required by looking at protein nanopore sequencing, for example with MspA:

The construction here is 1.3nm (nanometers), and it’s in a relatively high aspect ratio surface (literature suggests the sensing region is ~0.5nm). Even so, with MspA we see contributions from ~5 basepairs.
This can somewhat motivate us in terms of the feature sizes we’d like to see in a solid-state nanopore, certainly we wanting to be looking at single digit nanometer feature sizes.
In the semiconductor world, devices using a “5 nanometer” process are currently shipping. You’d be forgiven for thinking that standard processes can almost reach the required level of precision… well unfortunately, 5nm doesn’t mean 5nm. And while it’s relatively easy to create a coating only nanometers thick, the features in modern semiconductor are more like 30nm.
So standard semiconductor fabrication is almost 2 orders of magnitude larger what’s been shown to work for sequencing in biological nanopores.
As such, researchers have been going after novel fabrication processes. Using a variety of approaches they’ve achieved pore sizes of ~2nm:

Marija Drndićs lab does perhaps some of the more interesting work in solid state nanopore fabrication. The ~2nm pore in part (a) above comes from her lab and has feature sizes approaching those of MspA:
This publication shows traces from homopolymer single stranded DNA:
You can see that not only are the strands detected, but different homopolymers result in different current blockages. This is a great result, but as far as I’m aware about as far as we’ve been able to take solid-state ionic current nanopores.
The strands are unfortunately translocating far too quickly to be able to determine even coarse features of the sequence. Others have taken this work in a different direction adding labels to the strands. In particular Ulrich Keyser’s group has a bunch of interesting work using nanopipette for applications including DNA data storage.
But as far as I’m aware, no-one has shown anything approaching single base resolution on an ionic current solid-state nanopore and Marija Drndićs work seems to be moving toward a different sensing methodology.
Motion Control
If we can’t sense DNA at its native translocation rate it seems obvious that we would want to slow it down. And if we’re going to slow it down, it seems obvious to use the same method used with biological nanopores, enzymatic motion control.
Unfortunately as far as I can tell, there’s not much work on combining enzymatic motion control and solid-state pores. In one recent publication a group showed that the Phi29 polymerase was active when confined to a solid-state pore, has potentially a prelude to using this mechanism for motion control:
But it’s not clear to me why more effort hasn’t been made in this direction. Perhaps the work required to build this system isn’t considered worth it. One potential criticism is that the result would be a platform very similar to a biological nanopore sequencing platform, likely with poorer fidelity due to the pores larger feature sizes.
Other solid state motion control strategies have been investigated as well, for example Armonia with their use of Tortuous nanopores, Nooma with their dual nanopore approach, or mechanical approaches . Buffer viscosity has also slowed translocation by ~2 orders of magnitude, but unfortunately this is still a few orders of magnitude faster than enzymatic motion control.
Summary
Ionic current solid-state nanopore sequencing has been pursued in both the academic and commercial space. Notable commercial efforts include: Northshore, Nooma, and Nabsys.
However no-one has demonstrated a platform that approaches anything close to sequencing to my knowledge. And with biological nanopore sequencing on the market, the big question is would a solid-state ionic approach be any better?
Throughput is likely to be similar, but could we see an improvement in terms of either cost of accuracy? Unfortunately improvements here are also likely hard to sell.
While semiconductor nanopore arrays are not cheap, I suspect at least the manufacturing process uses standard substrates and largely established fabrication processes. Currently, solid-state nanopores don’t have this advantage. A low unit cost might be possible, but this is a difficult proposition if it means building a 100 million or billion dollar fabrication facility.
In terms of accuracy, it seems unlikely that you’d see much of an improvement unless much smaller feature sizes can be achieved. It’s possible however that the lower noise associated with solid-state pores could eventually give them a slight edge.
For all the potential issues, I still think it would be interesting to see a working demonstration of a solid-state ionic current nanopore with an enzymatic motion control system. In particular, it would be interesting to see if such a platform could be made easily washable (which is not doubt a difficult research problem in itself). If this resulted in a sequencing platform where the nanopore sensor was no longer a consumable, this could be very attractive…



An exciting proposition. Thanks for the introduction to the concept.