Olink
Olink is developing a novel sequencing based proteomics platform.
Their 2020 revenue was $54.1 million, 16.7% growth on 2019. This year growth looks set to be higher (in the 50% range). Interestingly the majority of their business seems to be service related (rather than kits). Margins seem to be ~60%.
Olink have a number of offerings, including a new instrument, the Olink Signature Q100. This appears to be an integrated qPCR platform. This doesn’t interest to me so much. I find their NGS based PEA approach more exciting as it lets you assay >1000 (different) proteins. The qPCR approach is limited to ~96 to 314.
In this post, I briefly review the approach and my initial thoughts.
The PEA approach
I’ve summarized the PEA (Proximity Extension Assay) approach in figure below.
Essentially Olink have designed pairs of antibodies with oligo nucleotide tags. Each antibody pair binds to a single protein (type). The antibody pairs are designed with a complementary region. These regions hybridize. The hybridized region is used as the primer for extension by a polymerase resulting in a longer fragment which embeds the complementary region:
So, in the ideal scenario, you only get extension products when both antibodies bind to the target protein. This should help reduce the false positive rate, as you need two concurrent binding events to get a signal.
As it goes that sounds great, but there are a number of potential failure modes:
In particular, we could imagine that there will be some background signal just from antibodies free in solution. They will stochastically encounter each other, the tags will hybridize, and be extended.
The second point is that antibodies are pretty big compared to target proteins. Schematics generally show them as tiny things, but they are likely several times larger than the target protein. This means that antibody pairs probably need to be carefully selected such that both antibodies can bind at the same time.
Finally, hybridization isn’t 100% specific. So lets say we have an unpaired antibody that shows weak binding, the oligo tags while not perfectly complementary, hybridize. Extension occurs and a signal is produced. Many approaches that use hybridization are problematic for this reason. But here it seems like less of an issue. The sequencing readout should provide enough information to filter out these sequences. Non-complementary region may also be providing information about the antibody and can be used to filter non-matching pairs.
However overall it’s clear that antibodies and oligos must be carefully selected to avoid issues. I imagine the development of these antibody libraries must have been a significant effort for Olink. Which makes the development of a library of 1000s of antibodies all the more impressive.
It’s important to note that we’re only identifying protein types, and likely don’t have a route to detecting single amino acid changes. For this reason it doesn’t make sense to directly compare Olink to next-gen protein sequencing approaches.
However Olink’s approach does seem like it would compete against a platform like Nautilus’ at protein fingerprints rather than full sequences.
It also competes against proposed protein sequencing platforms like QuantumSi, if we assume that they never reach their goal of producing full protein sequences, and in reality also only really generate fingerprints.
PEA-NGS (Olink Explore)
It’s fairly easy to see how the above scheme would work both with qPCR and sequencing. In the qPCR approach I assume the extension products have primer sites for amplification. In the sequencing approach, you just need to sequence through the extension product.
The sequencing approach (Olink Explore) seems to be a little less well developed than the qPCR platform. But there are several interesting recent papers on the approach. In particular, I’ve been looking at a recent Nature Communications article.
Correlation between NGS and qPCR for selected proteins looks pretty good:
It would be interesting to look at raw data across all proteins, particularly low abundance proteins to see where things break. However, raw data isn’t available “due to patient consent and confidentiality agreements”. On their website Olink state that “some proteins have a low correlation. Proteins with lower correlations tended to have limited spread in at least one platform and/or were typically close to the limit of detection”. But I don’t see a raw dataset anywhere.
Olink Explore seems to be available as a kit to use with your own sequencer now. This could mean that raw reads from this platform will start appearing. This would be interesting, as it should be possible to diagnose the various failure modes present in the raw data.
But the fact that qPCR and NGS correlate well is encouraging.
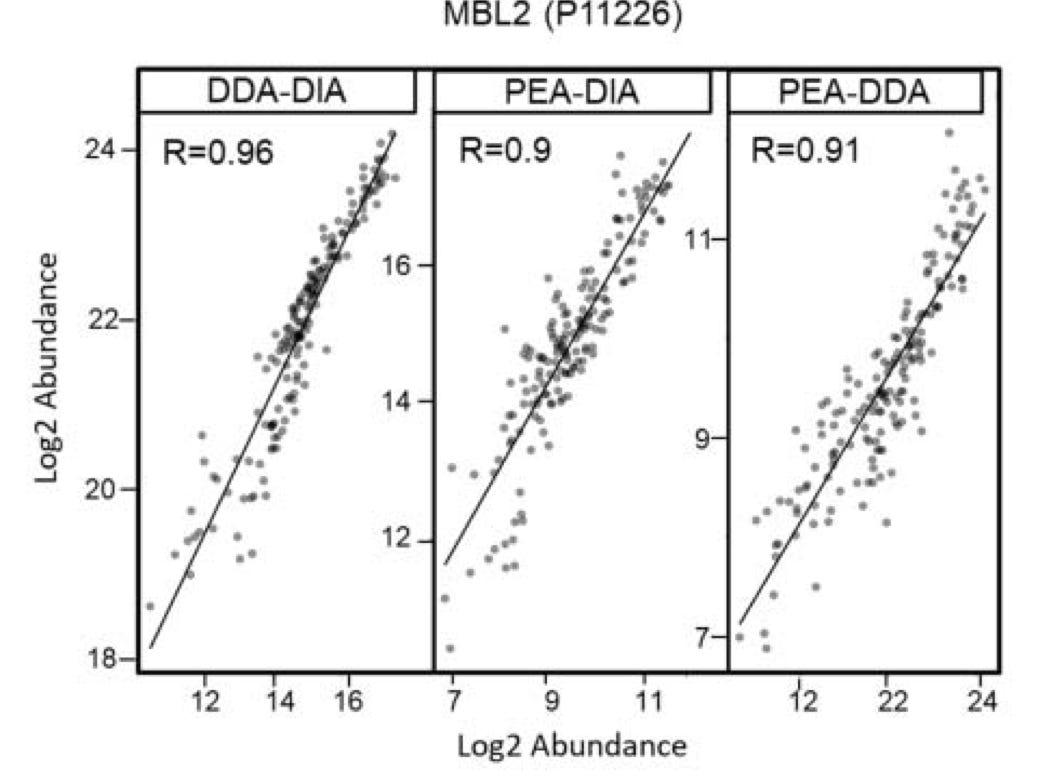
Other studies, have looked at how well the PEA/Olink approach is correlated with other approaches (below Mass Spec DIA and DDA):
They seem to correlate well in at least some cases, but the data I’ve found it quite limited. One of the issues with these kinds of validation studies is the lack of dynamic range in existing approaches, limits your ability to accurately assess a new platform.
Conclusion
Overall the Olink PEA approach is a neat idea. The data suggests that it works, and the financials suggest that there’s a market for the approach. I suspect that the potential failure modes ultimately limit the platforms accuracy. However, it’s not clear if this is fundamentally better or worse than other approaches.
Ultimately, coming from sequencing, I’d love to see a to see a high throughput single amino acid resolution sequencing platform. Olink certainly isn’t this. But it’s not clear that such a platform will be available in the medium term (say 10 years).
Perhaps Olink-type solutions are the best we can except in the short term.