Reticula Pt. 2 - COVID19 and Sequencing
In my previous post I discussed the background to Reticula and all the fun I’ve had trying to fundraise. In this post I wanted to discuss COVID19 and sequencing in relation to Reticula. Hopefully this is of some general interest, but if not, I promise there will be at most more one post on Reticula.
I want to start with the the failure of sequencing to make an impact on COVID19 diagnostics, and how that motivated the Reticula play.
Firstly, next-gen sequencing obviously had an impact on the initial sequencing of SARS-CoV-2 and variant monitoring. But everything I’ve heard suggests that even the most ambitious labs only sequence roughly 10% of the positive samples and many just pool samples for variant detection.
Various companies have tried using sequencing COVID19 for diagnostics directly. But none of these tests have gained traction. All together they probably have <1% market share based on the reports I’ve seen:
So why is that? Well… I don’t think it’s because sequencing is worse than qPCR. In fact, I think sequencing is probably better than qPCR in nearly every respect. And if we just sequenced total RNA, we wouldn’t see issues like tests failing with new variants. But the key to sequencing’s failure is that it’s too expensive:
I’ve seen various estimates of Illumina’s margins, but they seem to fall in the 70 to 90% range. For a diagnostic tool, my personal feeling is that you don’t want to multiplex 100s to 1000s of samples. Multiplexing adds complexity and cost, and I don’t think works for COVID19 diagnostics. So, if we look at Illumina’s cheapest run cost, that’s ~$500. Let’s assume the cost of goods is $50.
$50 is way too expensive for a COVID19 test. Again, if we use multiplexing we can probably get the cost lower… but the reality is that adds other costs and is undesirable anyway.
The Reticula play is therefore to get single run sequencing to $1 COGS on a $500 instrument (near the same price point as qPCR). The data quality is lower than Illumina sequencing, but the central thesis is that nearly any sequencing is better than qPCR. We’re not competing with Illumina… we’re trying to displace qPCR.
Now, a fair comment that is point is “but do clinicians even want this”. This is a reasonable objection. Clinicians possibly don’t want more than the “yes/no” answer that qPCR provides.
But I want it. And I think consumers in general want it. Because we don’t want another pandemic like this one. And without new tools, this is likely to happen again.
There are however other advantages with sequencing. In particular, because Reticula is direct RNA sequencing, our workflow should be simpler than both Illumina sequencing and qPCR and more robust to variants:
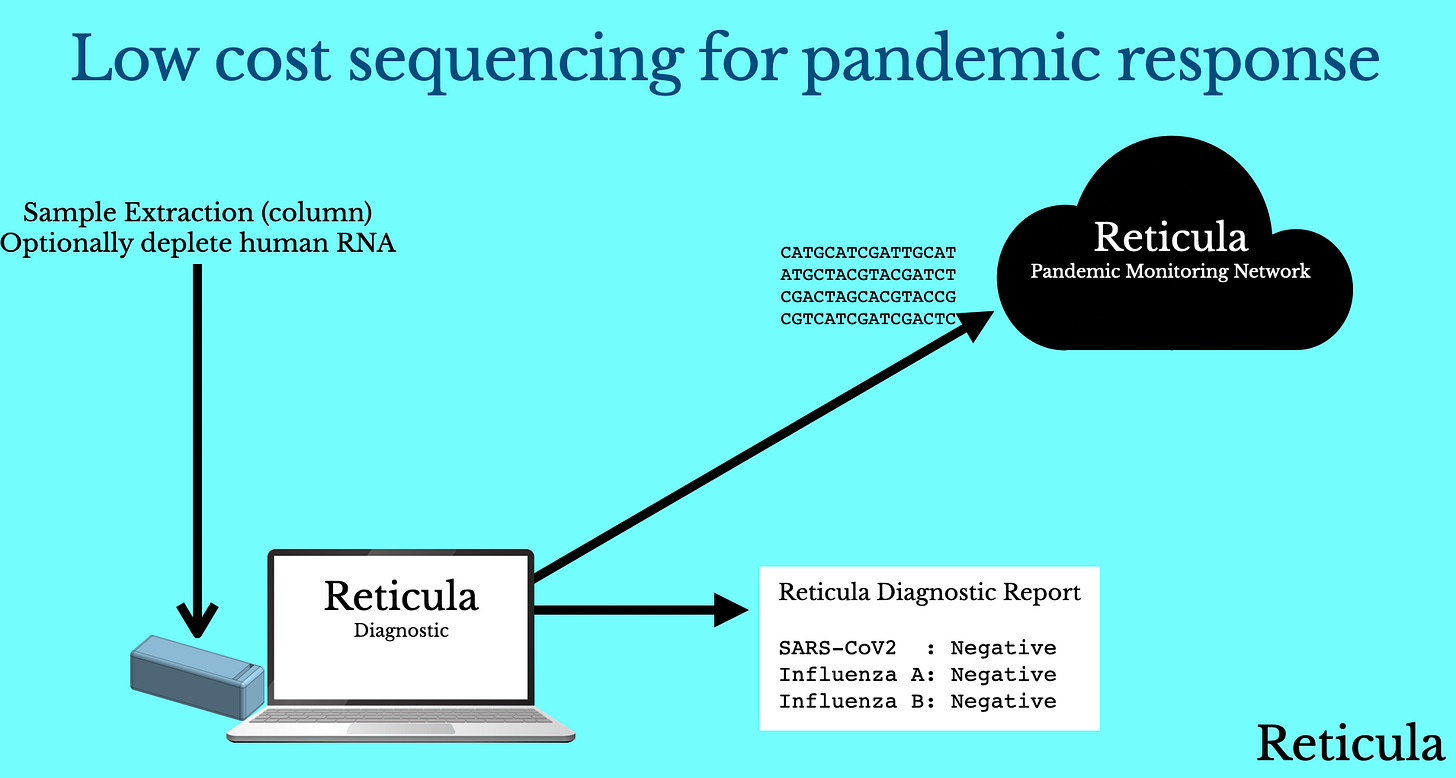
So, if we imagine a world where we have a low cost sequencing instrument. This instrument allows us to sequence a single sample, in less than an hour for $1 (COGS) on a $500 box, at a cost to the user similar to qPCR. How would we use this for pandemic response? There are two main phases where I think this is interesting.
The first is the one we’ve already discussed, use during a pandemic for accurate diagnostics (robust to viral evolution) and variant monitoring. But the second is the use of sequencing pre-pandemic:
What if sequencing based diagnostics had been routine around Wuhan prior to COVID19? What if people routinely took a diagnostic test when they had a fever (I don’t know about elsewhere, but it’s common in Japan to get a Flu test when you have a fever).
Now again, I agree, I’m not sure clinicians what to provide this level of feedback. But as a consumer… I kind of want it…
So with sequencing cheap enough to use routinely, users take a sequencing based diagnostic when they show symptoms. They get feedback on their test results. But… what’s potentially more interesting is what happens when there’s unknown RNA in the mix.
Potentially, and obviously this is another regulatory issue, this unknown RNA sequence could get uploaded to a cloud monitoring network. This would give us advance warning of a new viral strain in circulation.
What if this had happened, in Wuhan pre-November 2019 when COVID19 likely first appeared? How would this have changed the course of the pandemic?
My thesis is that this would have made a major impact on the progress of COVID19. It took us nearly 2 months to get the initial sequence of SARS-CoV-2, with universal sequencing we could have had this near day 1.
This lead up time delayed the development of vaccines and diagnostic tests. It then took us nearly 4 months to get just 1 million tests performed in the US. We just didn’t have the kits available early in the pandemic, they needed to be designed, approved and produced because each new viral test is specific to a particular virus (in this case, to SARS-CoV-2).
With a sequencing based test this wouldn’t be the case. If you sequence total RNA (and there’s enough viral RNA present in saliva that you should see it if you sequence, say 1 million reads) your diagnostic is virus agnostic.
This means that sequencing kits can be produced ahead of time, and stockpiled as part of a pandemic preparedness program. When a pandemic occurs you just need to update your software with a new viral sequence to allow reporting.
This requires a complete rethinking of how we do viral diagnostics, and again I think there are regulatory issues. But I don’t want to see another pandemic have the impact that COVID19 has, and as someone who works in sequencing, a pandemic where next-gen sequencing has really had a quite limited impact on the course of the pandemic.
By this point I’ve, hopefully outlined why I think cheap sequencing is important for this and future pandemics. Briefly let’s discuss exactly how good the sequencing needs to be. Accepting at nearly any sequencing is better than qPCR, what are our minimal requirements.
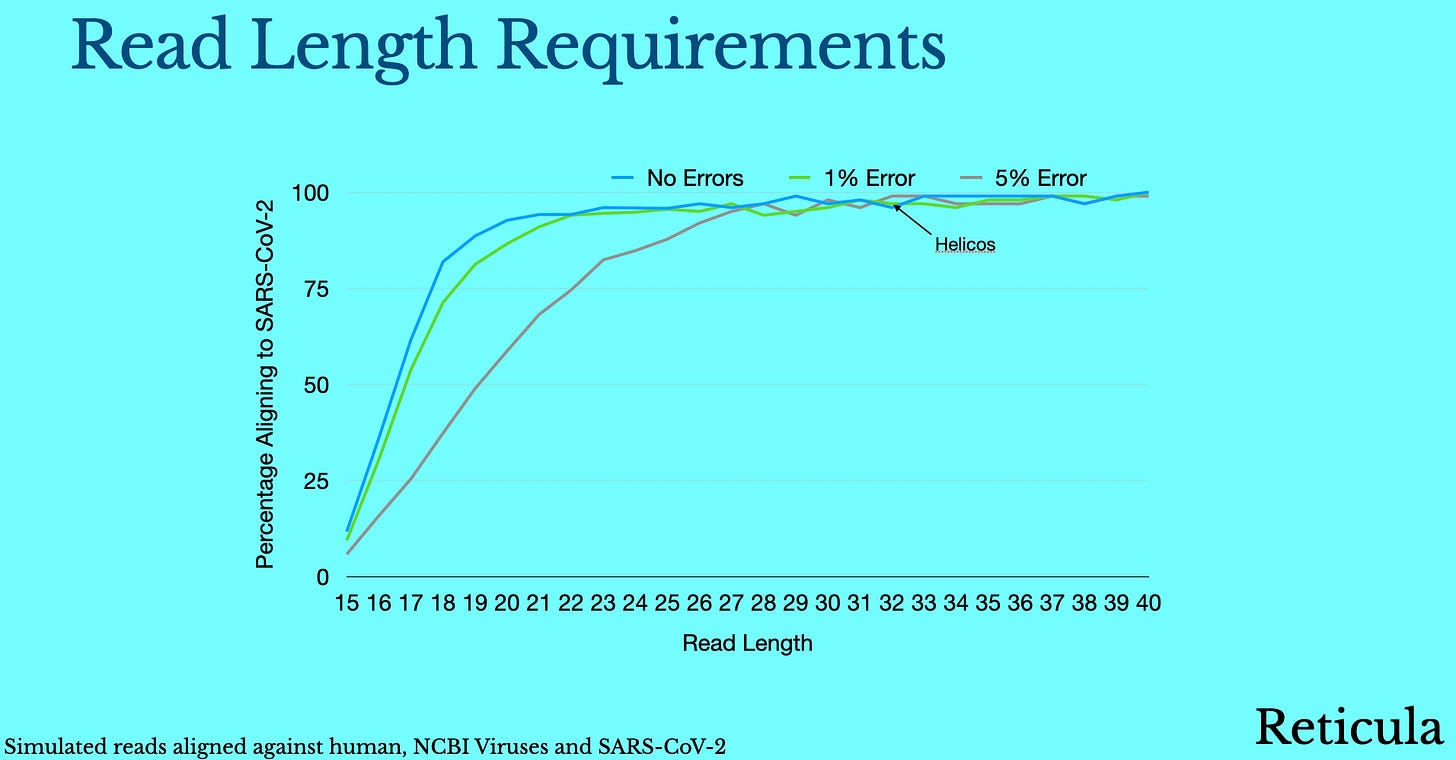
We’ll it turns out they are pretty modest. We can actually specifically align reads to SARS-CoV-2 even if they are as short as 15bp. If you simulate random human reads and apply random errors, they’ll almost never preferentially match against human. So a true single SARS-CoV-2 read of 15bp with 5% error rate, may fail to align against SARS-CoV-2, but a random or human read will almost never result in a false positive.
This means our false positive rate can be very low.
Now, on the true positive side. We can to an extent make up for errors with throughput. Even with 15bp reads and 5% errors we some percentage of true SARS-CoV-2 reads will align to SARS-CoV-2 allowing detection.
These are the absolute minimum requirements >15bp reads, <5% error rate. But we’d be in a much more comfortable place if we could do better than that (noting that we need to be $1 per run to compete with qPCR and that we can’t compromise on cost).
The sweet spot is probably at some longer read length. When we get up to 25bp we can comfortably deal with viral (and even human) genomes. So our sweet spot is probably 25bp+ reads <5% error rate:
This was the initial idea for Reticula. Focus on low sequencing run cost above all else to address applications current sequencing approaches can’t. And hopefully have an impact on this and future pandemics.
In the next post (if there is one) I’ll detail a low technical risk approach to achieving this.