RH Genetech
While looking into Chinese nanopore companies I came across RH Genetech. The company appears to sell a number of instruments including air sampling LAMP devices. However, I’m mostly interested in their two solid state nanopore platforms “FastVirus” and “AcidOn”:
FastVirus
FastVirus appears to use large (60 to 200nm) pores to detect whole viral particles. This sounds somewhat similar to Japanese startup Aipore, which is based around this publication.
I’m not a huge fan of this approach. SAR-CoV-2 varies in size from 70 to 90nm. This is on the same order as, for example, exosomes in Saliva. As such, the particle size is not very informative. Your hope then is that proteins on the surface of the virus will give some kind of characteristic signal.
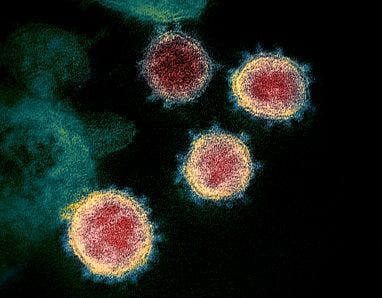
TEM images of SARS-CoV-2 highlight the problem with this, the spike protein is ~10nm, and not evenly distributed over the surface of the virus:
To detect features on the spike protein itself you are likely looking at needing nanometer to sub-nanometer resolution (similar to a nanopore DNA sequencing platform).
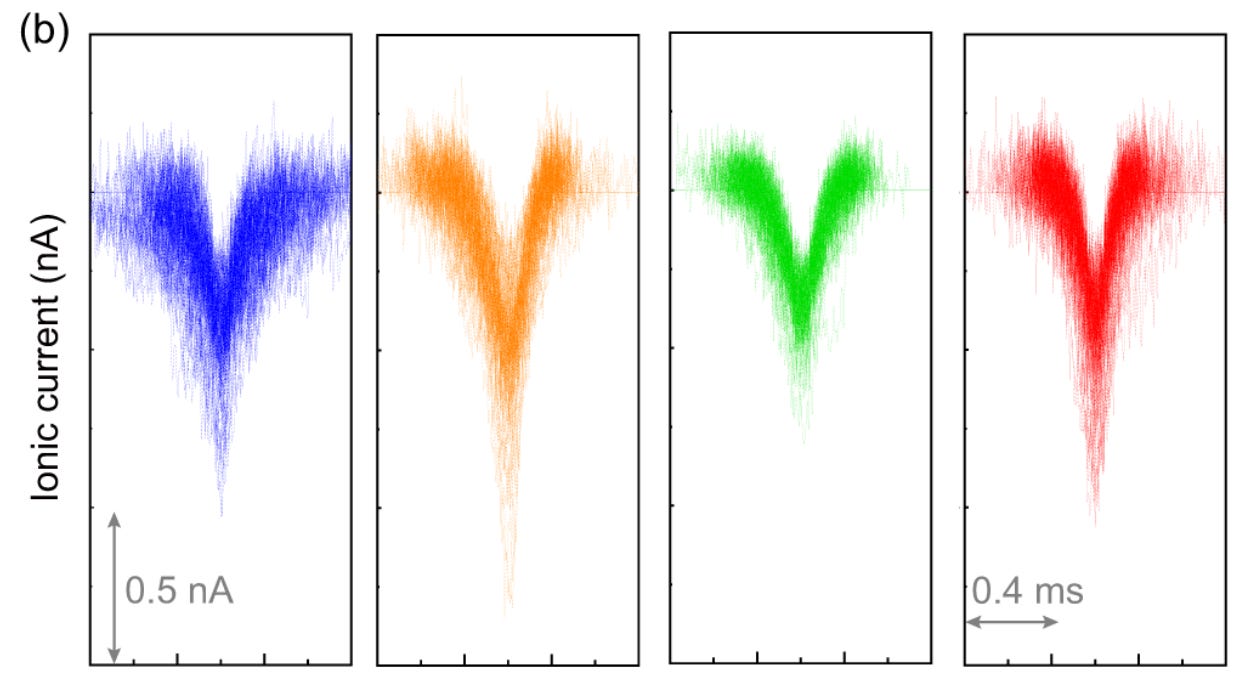
These particles will be translocating extremely quickly. The publication suggests ~0.5ms. They used a 1MHz sample rate, theoretically this could give a few hundred data points per spike protein. In reality the traces look noisier than this:
A larger pore, will naturally be noisier than a smaller one. You can achieve <10pA of noise on a protein nanopore. These pores show >>100pA. Nucleotides often show differences in current of <10pA. These are likely on the same order as features on the spike protein. Detecting a 10pA change on 100pA of noise on these traces seems very difficult.
On top of all this, unlike Oxford Nanopore’s approach, you don’t have any mechanism for slowing the virus down to make them easier to detect.
Overall, it seems unlikely that you will be able to identify any features on the spike protein at these speeds and with this much noise.
Both AIpore and RH Genetech seem to be proposing that the magic of AI will solve these issues. Perhaps there are other surface differences that an AI can detect and allow viruses to be classified. I have my reservations…
AcidOn
AcidOn is RH Genetech’s sequencing platform. They state the platform has 2nm pores, 100,000 pores in 6 layers and a throughput of 10Gb/h. Nucleotides translocate at a speed of 20 bases/ms.
20 bases/ms is crazy fast, and perhaps this is a mistranslation. Certainly 20ms per base would make more sense. This would require some method of slowing down the translocation speed (perhaps enzymaticly) . 2nm is still “big” by protein nanopore standards, but it seems vaguely possible that it could be made to work if they have a method of slowly the translocation speed.
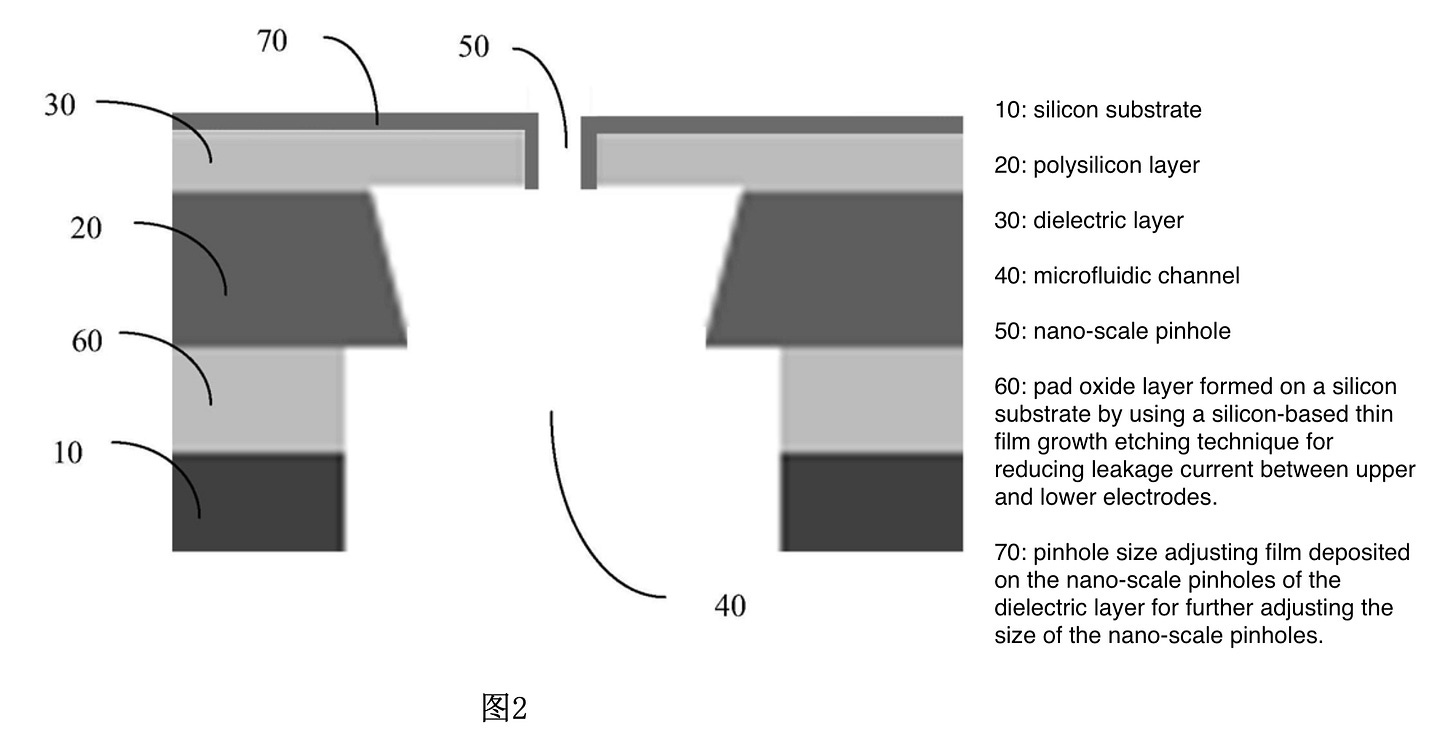
A quick patent search only revealed a patent relating to the pore construction methodology. This patent contains a schematic of the pore which probably tells you most of what you need to know:
What they seem to describe is a “tunable” nanopore. That is a pore which you construct and then fill in to further adjust the size. This sounds a lot like Northshore Bio (previous blog post). As far as I can tell Northshore are no longer around, I don’t recommend trying to visit their site at work as it now redirects to a Chinese porn site…
While I’m not an expert on solid-state pores, this approach doesn’t seem super exciting to me. The patent suggests that they’re looking at ionic current detection. This means that the length of the pore is also critical. So while it seems like the approach might make the aperture size smaller, it will increase the layer thickness…
Summary
Overall based on what’s been disclosed neither FastVirus or AcidOn seem to have novelty over approaches that have been tried by others. Of course it’s possible that there’s other IP that hasn’t yet been disclosed. But, based the basic physical limitations of the approaches described above, I suspect they will be tough to get working.
But who knows, there seems to be a surprising amount of nanopore development work happening in China, and I suspect they have resources to keep pushing forward.
Next time I’ll be looking at another Chinese nanopore startup, which seems much pragmatic. If that seems like it might be of interest, please consider signing up for the free newsletter.
After this, there will likely be a summary post for paid subscribers.