Thoughts On Pacific Biosciences
Let’s talk about PacBio (single molecule sequencing)!
Here are my headline thoughts:
PacBio raw accuracy has only increased modestly over 4 generations of instruments.
There therefore appear to be fundamental factors limiting raw read accuracy.
Raw read length has been steadily increasing.
PacBio made the marketing decision to trade read length for accuracy using the cyclic consensus sequencing approach.
For the more patient among you, let’s unpack and justify these statements.
Raw Sub-read (CLR) Accuracy Hasn’t Improved Much
After a quick search I found the following data on PacBio. While incomplete1 it’s clear that accuracy has only shown modest improvement at the sub-read level going from ~13% on the RS II to ~10% on the Revio.
The sources for this table are over on the blog.
Fundamental Limitations?
The accuracy could be fundamentally limited by polymerase kinetics…
I’ve tried to describe the various possible error modes in the schematic below:
One of these (and perhaps it seems the most significant) is what PacBio patents describe as the “branching fraction”.
The branching fraction is define as “a relative measure of the number of times a correctly paired base … leaves the active site of the polymerase without forming a phosphodiester bond”.
That is, the correct nucleotide sits in the polymerase active site (and therefore is detected) but doesn’t incorporate and just leaves. This will cause an insertion error2.
Understanding how these errors break down is more difficult. But some of PacBio’s polymerase engineering patents describe their efforts toward reducing branching fraction in particular.
In this patent they describe Phi29 mutants with branching fractions down to ~6.5%.
This patent is from 2012 but as accuracy seems to have improved only modestly since then, I think it’s reasonable to assume that branching remains a significant issue accounting for likely >50% of errors.
At the same time, the imaging system likely has improved signfiicantly. But this doesn’t seem to have helped improve accuracy much... My guess is therefore that accuracy is limited by polymerase kinetics. And perhaps this is difficult to avoid3.
But read length has been getting better!
One of PacBio’s very early patents talks about the effect of illumination on read length. They make a strong case here that photo-damage limits read length. Here they perform a number of experiments which support idea that an excited fluorophor in the active site of the polymerase is what causes damage to the strand under synthesis (not just laser illumination in general) and terminates synthesis:
As such, they’ve made significant efforts to protect the incorporation site:
Various methods for removing O2 (in addition to regular oxygen scavengers).
Shield Proteins which protect the active site.
It also seems likely that improvements to the optical system (like moving to a chip based platform have also reduced the amount of illumination required.
All these approaches have likely helped reduce photo-damage and increase read length from 1Kb on early systems to >160Kb4 on the Revio.
But Nobody Wants (very) Long Reads!
However PacBio seem to have decided that nobody wants 160Kb reads if that comes at the cost of a 10% error rate5. They’re probably right and they likely have the sales data to show this.
As such they’ve been using the CCS (and HiFi) approach to trade read length for accuracy. Going so far as to completely deprecate CLR (single long reads) on the Revio.
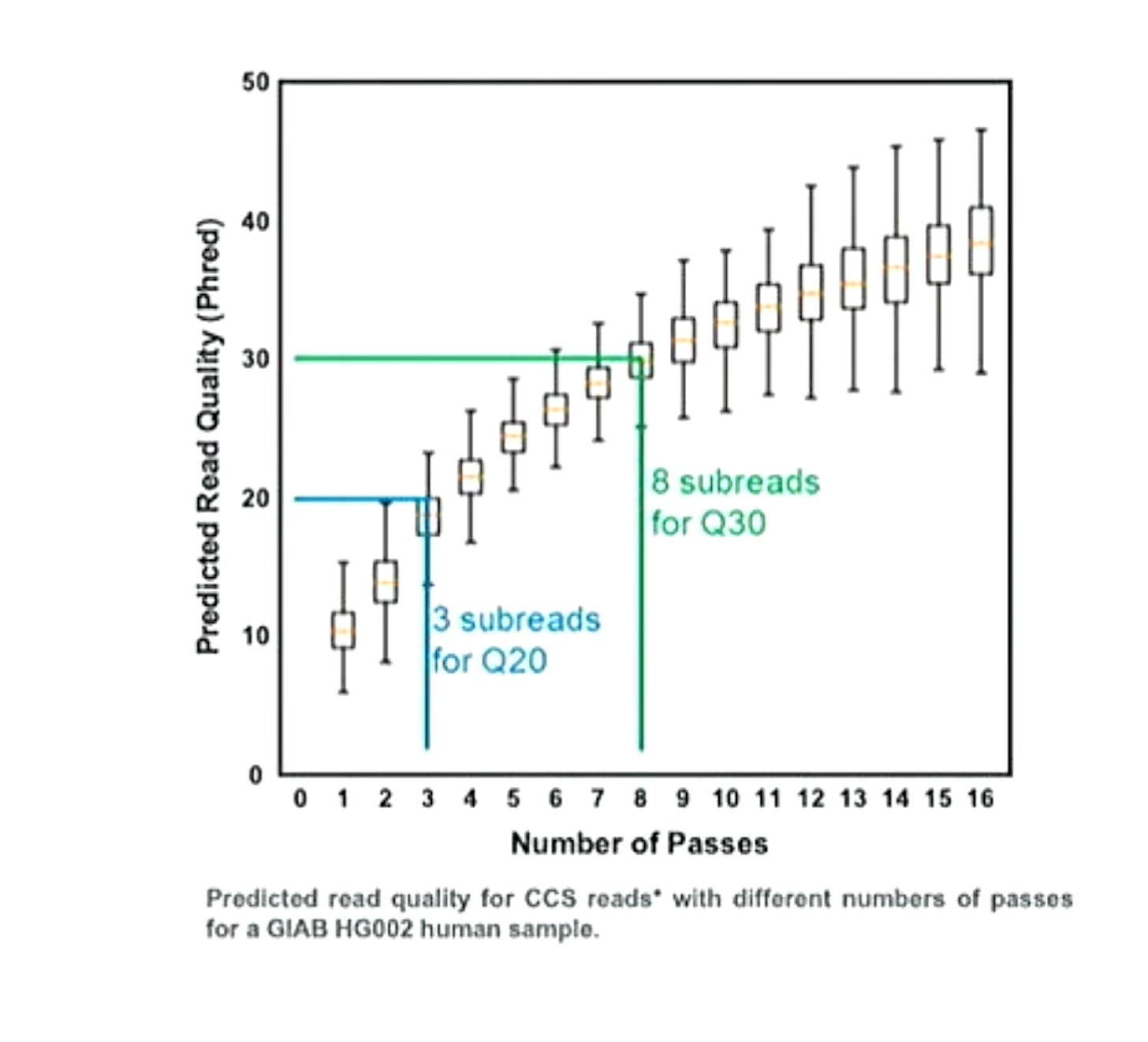
Helpfully PacBio have shown subreads versus accuracy plots for the Revio6:
With their read length improvements PacBio have been probably been able to increase the fraction of CCS reads that have >8 subreads at ~20Kb. So it appears that they made the marketing decision to deliver only these reads to users, where they can achieve Q30 accuracy.
Ultimately this seems like a solid decision. I’d take Q30 20Kb reads over 100Kb Q10 reads for most applications (particularly in human genetics).
As some have suggested you can make a strong case for Q307 20Kb reads being the new “clinical standard whole genome” giving you the ability to sequencing through many repeat expansions while retaining accuracy on SNVs.
And I imagine PacBio feel the same way.
Think i’m right? Think I’m wrong? Total nonsense? Reach out! Either by email (new@sgenomics.org) or on the Discord.
please forward additional references if you have them!
This will be a homopolymer error. Single A called as AA etc. and it would be interesting to dig through raw datasets and see what fraction of insertions are homopolymers errors.
You can imagine that if you try and reduce branching you might reduce affinity for the correct base in general and therefore your correct nucleotide branching fraction goes down but your incorrect base detection/incorporation rate goes up?
if you concatenate all sub-reads.
And as someone noted on Discord, there’s not much that gives PacBio CLR reads an edge over Oxford Nanopore (which can generate longer reads and on a cheaper instrument).
At least this is the assumption as this talk is all about the Revio… it’s possible they decided to use a Sequel II dataset? but I would hope unlikely. For reference the Sequel II also seems to require 8 subreads to reach Q30, further supporting the notion that raw read accuracy has not changed between the Sequel II and Revio.
And as far as I can tell, here we’re looking at emQ per-base Q30 accuracy not per-read modal accuracy or some other metric that is difficult to compare with Illumina or reason about…