Why Does Cluster Generation Take 5HOURS!?
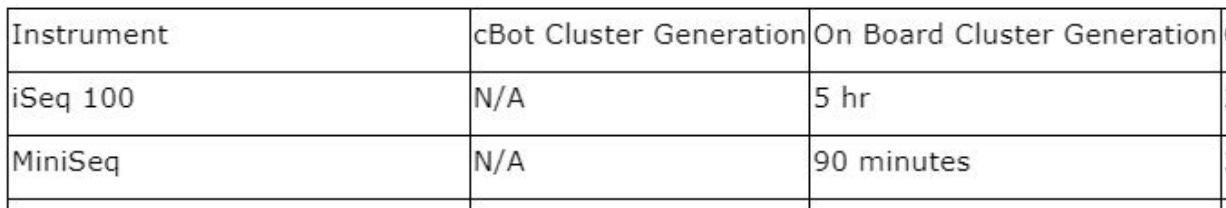
An interesting question came up on the Discord after my cycle time post. Why Illumina list such a SHOCKINGLY long time for iSeq Cluster generation:
5 HOURS! That’s 5 times as long as the MiSeq! But what’s weirder still is that cluster generation time in general seems to be trending upward on ALL instruments. The HiSeq 2500 took 1.5h, NovaSeq 6000 ~2h and the NovaSeq X takes 4.6h!
There seems to be no clear correlation between well size, or the use of patterned flowcells.
What’s going on?!
While it clearly adversely affects runtime something seems to be pushing Illumina to increase cluster generation time. Let’s try and figure out what it is…
Here’s my best guess: Illumina are doing everything they can to increase cluster template density.
Higher density means they can move to smaller clusters and decrease imaging time. But likely comes at the expense of cluster generation time.
Cluster generation in the original patterned flowcell patents took about an hour:
“patterned flow cell at 38° C. Incubation was continued for 1 hour at 38° C. before washing with HT2 wash buffer (Illumina, Inc., San Diego Calif.) and SyBr Green staining the clusters. Clusters were then processed for sequencing by LMX1 treatment for 30 minutes to linearize the DNA in the clusters,”
Similarly back in the original Isothermal bridge amplification patents a 1 hour incubation time was used.
Let’s see what other techniques they’ve got going on... Well, patents suggest that the move to patterned flowcells helped push up primer density and template density in clusters:
“Surprisingly, for flowcells prepared as described above a 3 to 10 fold increase in primer density was observed over metal patches as compared to the density of primers over interstitial regions. Similarly, a 3 to 10 fold increase in signal intensity was observed for DNA clusters formed over metal patches compared to signal detected from interstitial regions”
Then in a second patent they discuss further methods for increasing template density:
“it has been surprisingly discovered that combining standard bridge or ExAmp surface amplification with cleavage of one of the primers, followed by a sideways boost, yields amplification product that is many times more robust, enabling significantly higher utilization of surface primers and generating clusters that are many times brighter during optical scanning analysis. It was unexpected that enhanced occupancy would result from cleavage of one of the bridge amplification primers.”
However this second approach seems like it would require a 3rd primer to enable paired end sequencing, this doesn’t currently seem to be implemented.
There’s another interesting note in this patent however:
“The term “amplifying” or “amplification” herein is intended to mean the process of increasing the number of a template polynucleotide sequence by producing copies of the template. The amplification process can be either exponential or linear, but is typically exponential. In exponential amplification, the number of copies made of the template polynucleotide sequence increases at an exponential rate. For example, in an ideal amplification reaction of 30 rounds, one copy of template DNA will yield 230 or 1,073,741,824 copies. However, bridging amplification as described herein does not typically occur under ideal conditions, and a 30 cycle “exponential” reaction may only yield a few hundred to a few thousand copies of the original template, mainly due to the limited localized concentration of surface bound primers and the competition with template rehybridization. In linear amplification the number of copies made of the template polynucleotide sequences increases at a linear rate. For example, in an ideal 4-hour linear amplification reaction with a copying rate of 2000 copies per minute, each copy of template DNA will yield 480,000 copies.”
Interesting… What else have we got?
Well there’s this, which uses a “non-sequencing primer” and is listed under NovaSeq X patents on the Illumina site:
“The sequencing primer may be used in binding and amplifying deoxyribonucleic acids (DNA) or ribonucleic acids (RNA), while the non-sequencing entity does not participate in binding or amplifying. Rather, the non-sequencing entity acts as a spacer between sequencing primers. Spacing out the sequencing primers may enhance amplification by limiting steric hindrance for proteins involved in the amplification process.”
But it didn’t seem like the results were very compelling or would result in increased cluster generation time.
I don’t have much else to go on and no hard answers. But my best guess is after the break!