Browsing through random sequencing company websites (as you do) I noticed that Qitan Technology have released a couple of datasets from their platform!
Under “Demo Data” on their website they have datasets for both E.Coli K12 and a human cell line. I decided to take a quick look at the E.Coli data, and run it through BEST. The timestamps indicate that these runs actually come from 2021, but I don’t think I’ve seen this data before! Anyway, I thought it might be fun to take a quick look and see how the other nanopore company is doing.
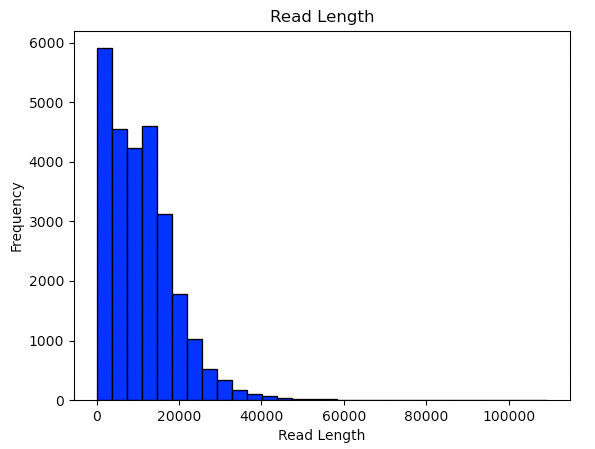
Firstly the read length distribution:
Nothing particularly special here, but it does appear to producing reasonably long reads!1. Doesn’t seem infeasible that they could push this out further with some optimization. Still at the moment it seems significantly shorter than shown in the best Oxford runs.
Next Q scores:
The Q scores seem quite poorly calibrated (here compared to the ONT Simplex dataset previously discussed). This data suggests they’re limited to Q25, but it’s entirely possible there are reference issues here (I grab a random K12 MG1655 reference). However… the overall error rate seems lower than I’ve seen suggested in Qitan publications previously.
The vast majority of bases are <Q15: