This was supposed to be a easy… just take a few snaps of the cameras in the HiSeq X and write up a quick post…
It turn into much more of a journey. It all starts with a HiSeq X camera. As regular readers will know I bought a HiSeq X and have be tearing it down.
The HiSeq’s use what’s called TDI imaging. You can think for it taking a picture by scanning across a surface a line at a time1. Kind of like a flatbed scanner!
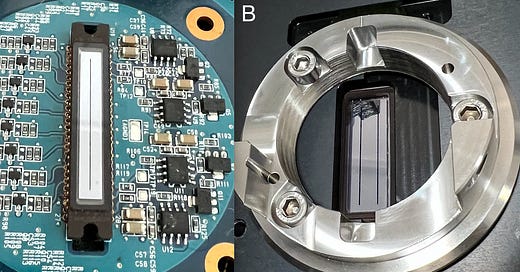
As such you only need a line sensors rather than a larger 2D camera. So you get these long thin CCD image sensors in TDI cameras:
The problem is the one I’d previously purchased (from a HiSeq 1000 or 2000) had a single “line”. The one in the HiSeq X had two…
What Hamamatsu have done (at Illumina’s behest) is fabricated two cameras on the same silicon substrate… so when you look at the HiSeq breadboard, it looks like there are two cameras. But actually there are 4… each “camera” contains two independent sensors and each unit has two separate cameralink outputs:
So we have a 4 camera system, suitable for use with a 4 color chemistry. Which is what you’d expect for the HiSeq X…
Each pair of cameras has two filters that sit right in front of the image sensor:
So I figured maybe they just illuminate the whole surface and then throw all the “green” emission at one pair of cameras and all the “red” at the other camera then filter it just before it hits the sensor.
This would be bad.
It would be bad because you’d be exposing the whole surface to light, not just the two thin lines where you are actually imaging the surface.
Flooding the whole surface with light would cause photo-damage to parts of the surface which are not being imaged. Ultimately this would effect accuracy and read length… and your DNA sequencer wouldn’t work as well as it could.
So they don’t do that.
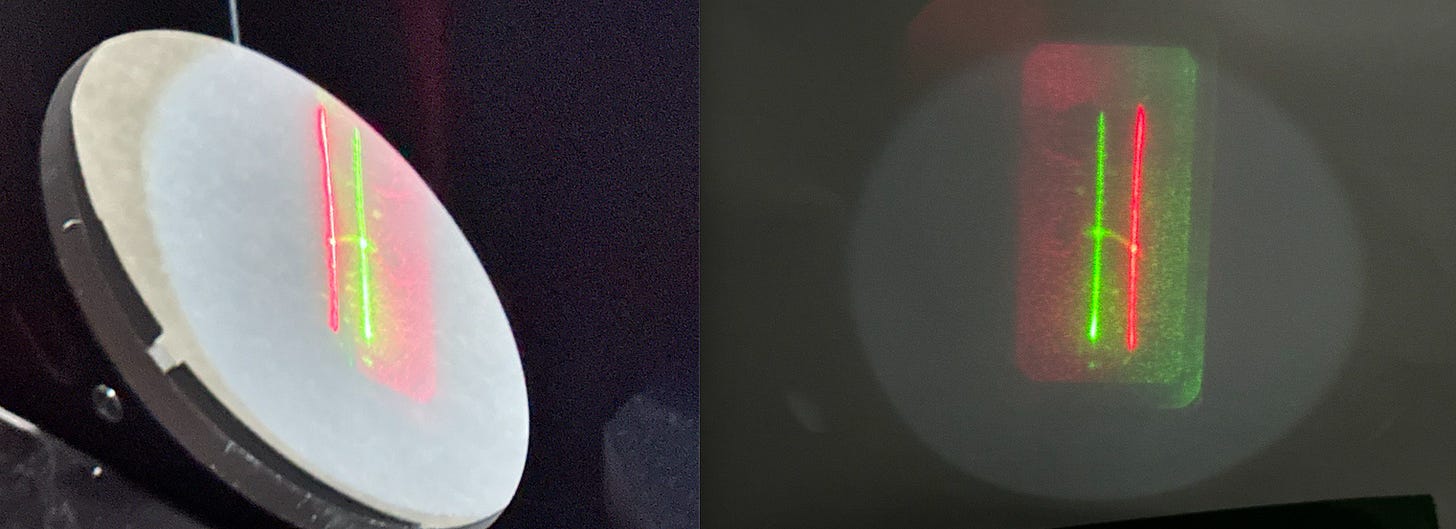
If we remove the camera, fire up the lasers and try and get everything focused on a sheet of paper we see a nice clean line2:
If we power up both lasers we see two lines offset from each other:
So here’s what I think is happening.