PacBio Revio Error Rate is <0.3%?!
Some kind users on the Discord have pointed out that the fastqs provided by ONT discussed in the previous post may also have been called using the fast model. Why the only fastqs provided in a public dataset would be called using a model that it seems is not recommended is unclear to me, but don’t worry! I’ll be finding some more ONT datasets to run through BEST!
However in the mean time I've thrown some PacBio Revio data through BEST to see what the statistics look like there! The Pangenome project has datasets for all major platforms (including some ONT duplex data!). So it seems this might be the best dataset to play with going forward! In any case I thought I’d quickly write this up while I’m at the beach and my daughter plays in the rain…
Dataset locations are in the footnote1 and I’m using the same reference noted previously.
I’ve aligned this using the same approach used for the ONT data (again with Dorado aligner, which may not be entirely fair to PacBio data).
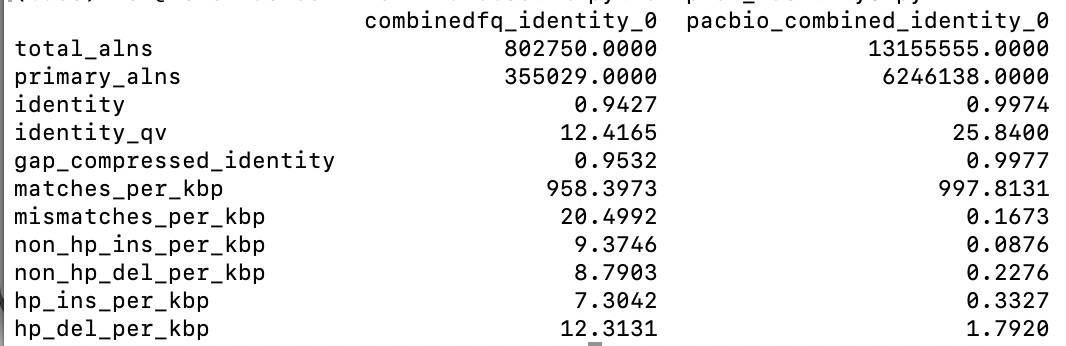
The empirical quality here is somewhat better than the Q scores they are assigning, see “pacbio_combined” in the plots below:
The population of basecalls above emQ20 also looks pretty solid!
And overall the identity is 99.74% putting the error rate somewhere around 0.26%:
Overall, for me this is close enough to the vendor specification (~Q30) that I wouldn’t be too concerned and it still feels like PacBio is currently providing the highest quality data of any current sequencing platform! With reasonable long, low error rate reads.
I’ll keep iterating over this analysis, there’s an Illumina run aligning at the moment. I’ll keep posting interim results, but some of those might now to go the new bitsofbio substack.
https://human-pangenomics.s3.amazonaws.com/index.html?prefix=submissions/80d00e88-7a92-46d8-88c7-48f1486e11ed--HG002_PACBIO_REVIO/
I used:
m84039_230117_233243_s1.hifi_reads.default.bam
The HG002 reference used is noted in a previous post.