A few of us from the Discord have been working on this DNA sequencing feature/pricelist. This is always a bit of a struggle, as some vendors can be less than explicit about pricing. But usually the platform specs are failure clear.
This isn’t the case for Oxford Nanopore’s instruments, where there has never been a clear throughput or accuracy specification. When I last went hunting on the SRA I estimated 72Gb a run from public datasets.
But recently Keith from the excellent OmicsOmics. Posted this on the Discord in response for my request for more run throughput information.
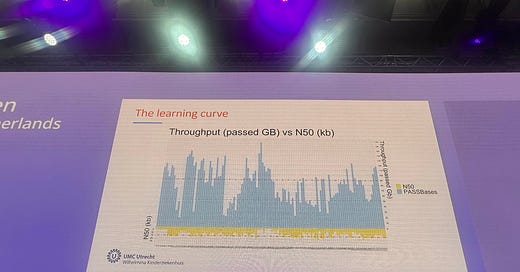
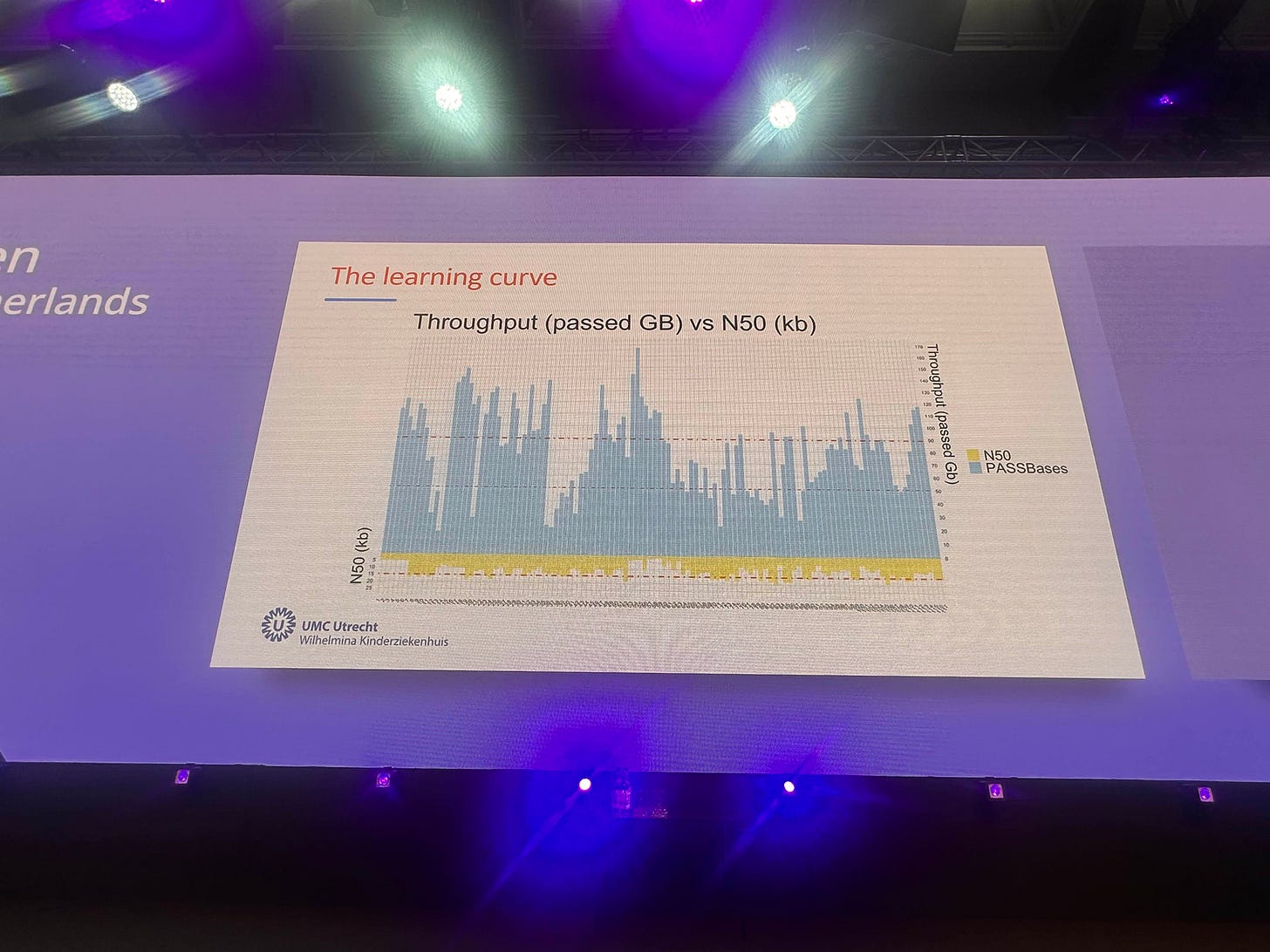
I went through and annotated1 the chart estimating the throughput for the 116 runs here, and calculating an average. This ended up being 84.06Gb/run. With only 13.68% or runs generating >120Gb of data2.
We of course don’t know exactly what filtering criteria was used here, but in general some very low quality reads are present in ONT datasets, which should be removed.
Another user on the Discord also posted this from PacBio Revio (SPRQ?) runs:
These are pooled runs with 16 libraries → 8 flow cells. Our understanding of the data is therefore that 87.5% of flow cells generated >120Gb of data here. That seems inline with the expected yield of 120Gb from PacBio. I’ve not calculated the average here, but it seems clear that it’s far higher than 120, possibly closer to 150? PacBio data is also pre-filtered on instrument, so this should all be high quality data.
In terms of the spreadsheet, I’ve stuck with 120Gb/run (the vendor spec) for PacBio) and 85.17Gb/run for the PromethION.
This puts PacBio ahead in terms of cost/Gb at $8.3 versus $10.6 (for a ProthMethION 32pack flowcell).
Beyond this, early data on PacBio flowcell reuse data was posted, suggesting 4x reuse generating 400Gb of data, which would push PacBio even further ahead.
As always, I welcome better data, come chat on the Discord.
Note, in the first version of this post I’d mislabeled a few of the data points, which a reader was sharp eyed enough to notice and point out. I’ve reviewed the svg correcting these, hence the throughput has been revised down from 85 to 84.




PromethION has an open front end where users run any kind of sample or prep. Without amplification in some cases. Therefore the front end is variable, that is a feature and comprises many of the customers. The only way they can provide specifications is by locking down the front end, homogenising the sample preps and fragment lengths, i.e. unpicking the main features that people buy the platform for. It is not difficult to run a library on this technology, then extrapolate how it is running from the first hour. If it isn’t on track the run can be stopped, flowcell flushed and a new library loaded so there is no risk/cost to having a open front end unlike other platforms where the run is committed once the sample is attached to the flowcell.