Yesterday we looked at a new nanopore protein sequencing startup, today we look at a label-free optical approach. This gives further credence to the idea that if you have a good new sensing technology you are now more likely to see that directed to Protein sequencing than DNA sequencing.
The company in question is PumpkinSeed. The work appears to be funded by the CZI, Gates Foundation, and Moore Foundation1 but it’s unclear what VC funding the company has received. And I assume they are seed stage.
There are a few bits and pieces floating around about the PumpkinSeed. Including this great Youtube video. There are also two published patents.
The second of these is a little more exciting to me as it discusses a label-free de novo protein sequencing approach. The approach is only briefly mentioned in their slides, but described in depth here.
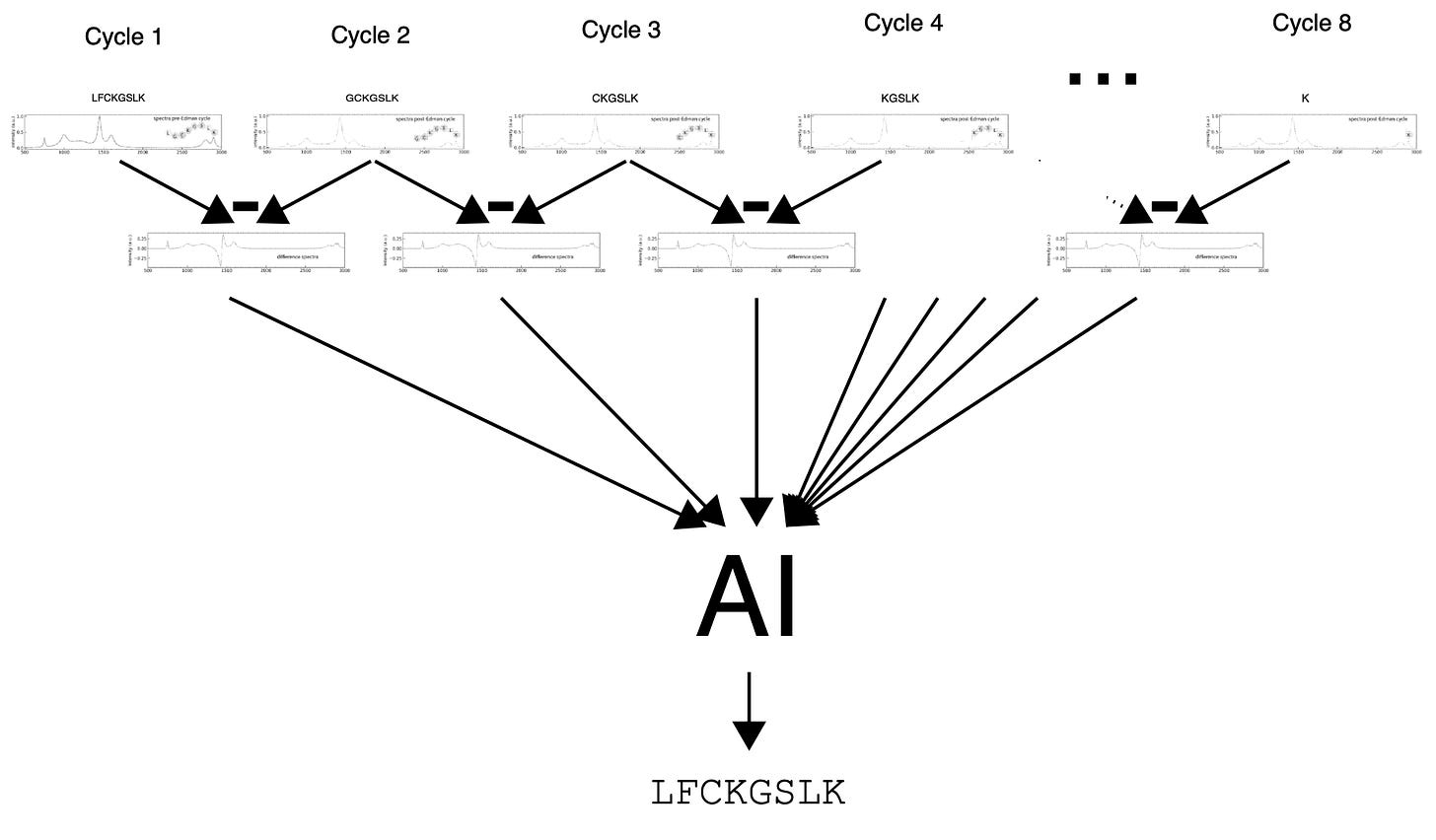
At a high level the approach is very simple. They take single molecule Raman spectra of short peptides. The terminal amino acid is cleaved, then another spectra is taken2 sequentially until you have Raman spectra after every cleaved amino acid.
Then you combine3 all the spectra of these incrementally shorter peptides to derive the original sequence:
Structures/Thoughts
This all sounds great in theory but some fancy tech is required to enable this. There’s a very nice YouTube video describing the academic nanostructure work which led of PumpkinSeed, I recommend checking it out.
Most of the work in the patent appears to be based on this describing “guided mode resonance structure, which can be configured to concentrate light”. This will help focus the excitation on a peptide of interest:
The Raman signal will then I assume need to go through a diffraction grating, and is projected onto an image sensor:

So far this seems reasonable, single molecule Raman spectroscopy is now a relativity well established technique. It seems reasonable that short peptides (8-mers in their examples) would provide sufficient information across multiple Edman cycles to identify the sequence uniquely.
I would have some concerns over the scalability of the design shown above. Projecting the spectra onto a camera would mean only a small number of peptides can be measured at once.
If sampling at a small number of discrete wavelengths is sufficient this may allow greater scalability. However they have the advantage over QuantumSi’s chip based system in that they can scan across a much larger flowcell to scale throughput. Given that QuantumSi is also dumping the chip, I suspect they wouldn’t dispute that.
Pending further details of the Raman spectra, I find the idea fairly interesting. It seems like it might be hard to scale much beyond 8 peptides to me4. That is likely sufficient for many applications, but could be the major limitation of the approach.
And like other protein sequencing approaches, the question remains “which applications” and “is the market sufficiently large”.
The patent shows a difference measurement between each cleavage cycle, I would hope that difference signal is somewhat indicative of the animo acid with a relatively simple base calling approach. However there’s a lot more information than just this in each trace, as you also have all subsequent amino acids in the signal. There are a few computational techniques which could leverage this information, but broadly “AI magic”:
Because you have increasing complexity/background… this feels like the main limiting factor…







Thanks for the nice summary Nava, very informative, especially the video. Does seem like a very hard scaling problem. Naive question: would the harsh treatments of Edman degradation cause single molecule sensitivity to be lost?
Regarding potential market size (IF technical hurdles are overcome) wouldn’t you expect NGPS to take over from Olink/Somalogic/Nomic etc? Given that Thermo paid 3B for Olink, presumably NGPS would command similar or greater numbers?
it's Raman, unless you're measuring noodles