Stream Genomics
Stream Genomics are working on a DNA sequencing platform, I’ve had a chance to talk to some of the folks there in the past and they seem nice. But we didn’t really talk about their technical approach.
There has been a fair amount of speculation over on the Discord. This had me trying to extract too much information from filenames briefly shown in the background of videos:
But now a patent has been published, so let’s take a look!
The patent describes a single molecule sequencing-by-synthesis approach, the closest thing out there would be Pacific Biosciences. Single molecules of DNA incorporate labelled nucleotides which are detected optically.
In order to confine excitation they use TIRF. Again this is very typical for single molecule imaging, having been used by Helicos and others.
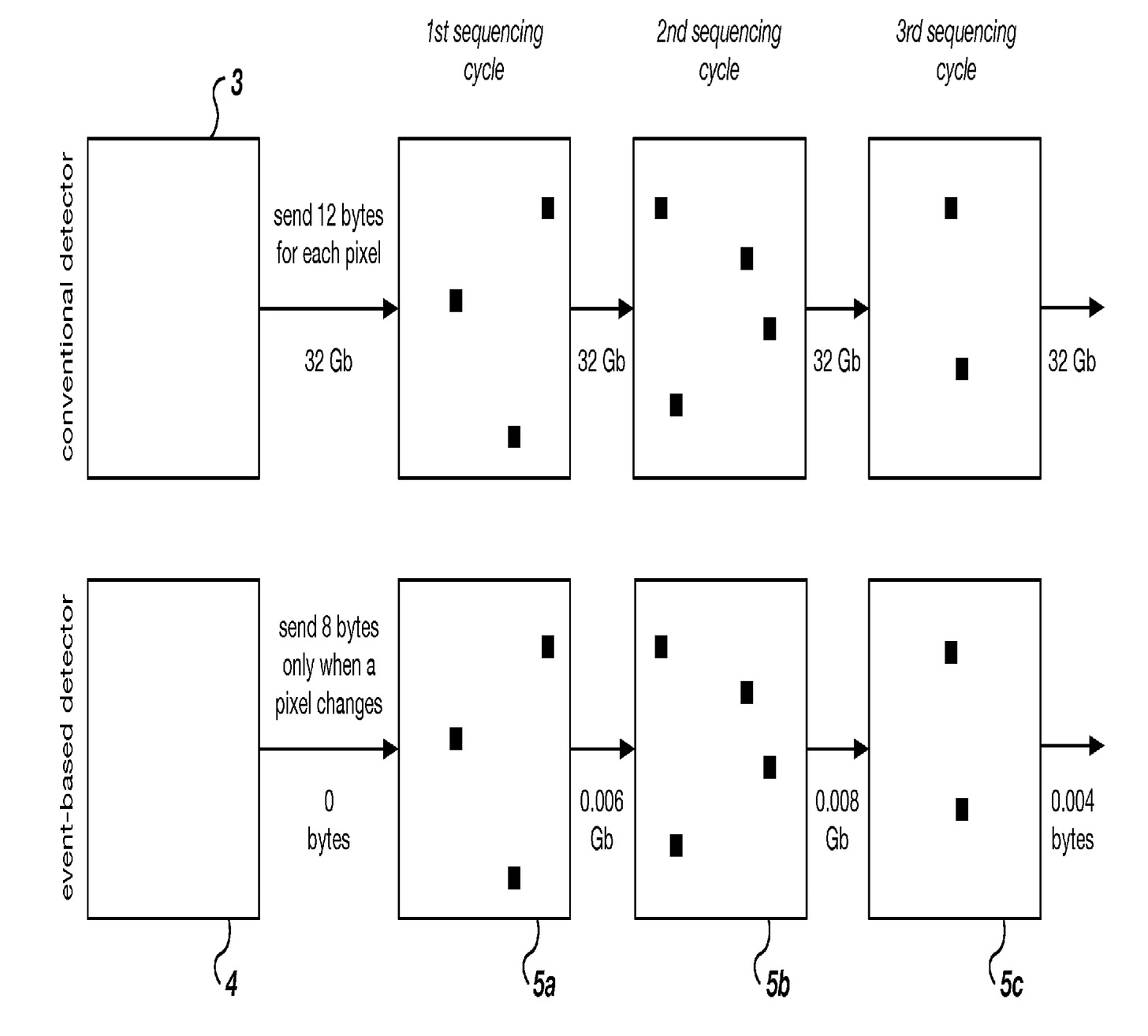
The innovation comes in the detector. Typically a sequencing platform will acquire images using a standard camera sensor. Just like a normal video camera these capture a fixed number of frames a second, typically 30fps.
This is true for PacBio too, their chips capture images at 100fps.
temporal contrast pixel arrays
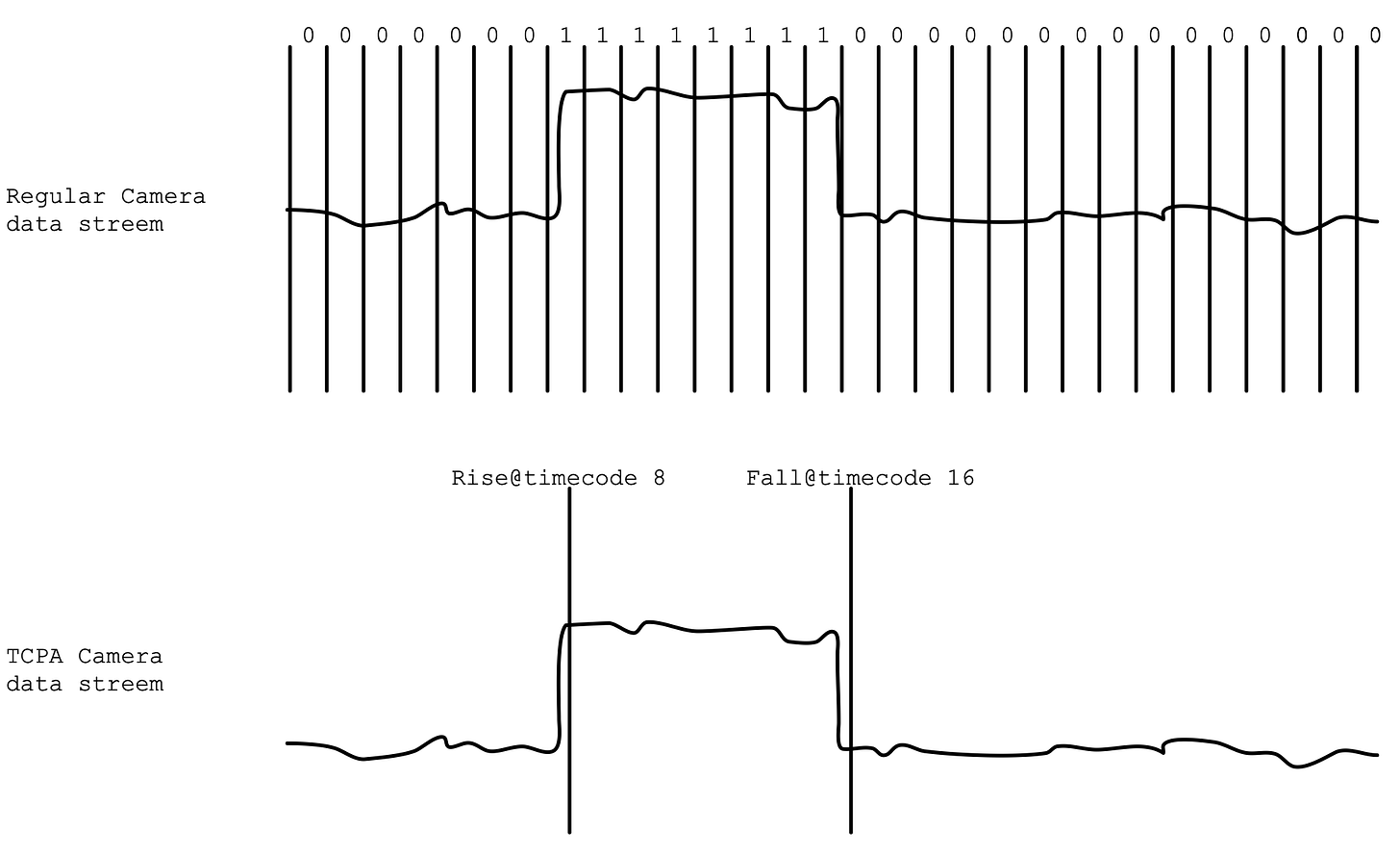
The fundamental innovation described in this patent is use for a new kind of sensor: temporal contrast pixel arrays (TCPAs).
The idea is that for each pixel, you only send information when the pixel value changes. Conceptually you might think about the data in contrast to a regular camera as follows1:
Obviously a TCPA data stream is much more efficient in terms of the quantity of data generated. You also lose a lot of information, you don’t get information about the variance of the signal within an event, sub-events etc.
Therefore the sequencing approach needs to be strongly coupled to the detection technology.
In order for this to be valuable you need to build it into the imaging sensor itself. A custom image sensor needs to be designed with analog electronics which does the rise/fall detection before passing the digital events over to an FPGA.
The potential benefits are two fold:
Less data may make it easier to engineer a much larger image sensor.
The downstream processing becomes less data intensive (cheaper FPGAs, base calling compute).
But the downside is that engineering a novel image sensor. Particularly one of the size required will be very costly. Manufacturing, in the relatively low volumes that DNA sequencers are sold is also unlikely to be “low cost”.
That said, PacBio can sell a novel image sensor as a consumable for $1000. So a few $1000s is likely tractable here. And for Stream the image sensor is not a disposable product, but part of the instrument.
So low cost consumables, moderately expensive instrument.
Examples
In the patent Stream give one example of a 20MP camera2 used to sequence a human genome. I don’t find this compelling enough, 43MP 100fps camera suitable for this application are readily available. An FPGA design built around these sensors which performs the desired data reduction would be nearly as compelling as the custom sensor approach described and far cheaper3.
To make the technical approach compelling I think you’d need to be pushing closer to 100MPs.
Summary
At a high level this patent seems to propose a PacBio-style sequencing approach with a few twists:
Rather than using a consumable chip, they use a regular microscope-style setup.
They use regular TIRF instead of ZMWs.
They don’t propose using CCS to correct errors/improve accuracy.
Without CCS their accuracy will be likely be very low. This may work for the applications they are interested in. Perhaps point-of-care like Sepsis (mentioned in the patent).
The problem is that the only real potential advantage TCPA gives you is scaling. And those applications don’t need scale. So it’s unclear why you wouldn’t just stick with a regular image sensor. Particular given the huge costs involved in developing and producing a novel sensor.
But, this is an early patent so it’s possible that both the target market and approach have shifted significantly. I look forward to hearing more from this interesting team!
Whole human genome sequencing is performed by an instrument with four large-format cameras with 16M or 20M pixels each. The instrument detects sequencing events during a time window of 10 to 100 milliseconds and reports changes that occur within an aperture of about 10 adjacent pixels (16-bits per pixel). The analytes are pipetted with a liquidhandling device into a 96-, 384-, or 1536-well plate and a stage is used to scan each well one at a time. The instrument detects at least 300 bases per minute. The platform can be capable of sequencing a whole human genome (WHG) at standard 30x depth of coverage in three hours. The reduced reagent quantities and efficiencies from analyzing only significant data results in reduced sequencing costs and improvement in speed.
Honestly I’d probably budget $10M+ for an initial run of smaller sensors. And then you have the problem that many novel sensors fall into. You need high volume to reach low cost per device. But without low cost (and even perhaps with) you can’t ramp to high volume.

Thanks for the writeup Nava.
I'm a bit puzzled how the chemistry will work without ZMWs (if indeed ZMWs are not being used). If you have ~100nm evanescent depth and ~300nmx300nm pixel, that gives a volume of ~0.01um^3 = 10^-20 m^3 = 10^-17L. If you have 1uM nucleotides then you'll have 10^-23 moles or 6 molecules in the volume at a given time, which will contribute quite a bit of noise. You can drop the nucleotide concentration of course but there's a speed cost to that. Unless the goal is to use synchronized reversibly blocked ncuelotides like ILMN wherein the excess can be washed out.
Pretty interesting nonetheless, thanks for surfacing!
Using TCPA may just solve one of the big problems with CMOS arrays. Since TCPA's only report when they detect signal, they run asynchronously so each spot basically reports at whatever rate it is running at. Now!!!! if they have a chemistry that runs continuously, so the reaction goes dye-ligated base binds emits signal and then the next dye-ligated base binds after ligation is completed (in other words) if all bases are present in mix and they have a way of displacing currently bound dye and adding the next base, then TCPA could just start reporting the sequence continuously as soon as rxn gmish is present.
Each spot would be continuosly reporting sequence.
See this amazing video about 20 years of PacBio development: https://vod.video.cornell.edu/media/2022%20CNF%2045th%2C%20Video%202%3B%20Dr.%20Mathieu%20Foquet%2C%20PacBio/1_tvaaaph6
I think I found this vide throu ASEQ. Was it was discussed in this channel. It is mind blowing!!! How much integration they achieved on a single chip!!!!