Ultima Genomics - More Reads Per Bead?
Summary: Illumina produces less than one read per well due to low PF rates. Could Ultima have an edge here by using a method that generates a read from every bead?
A Background On Polycolonity
You’ve got this problem in DNA sequencing… in order to produce enough signal you generally want to create multiple amplified copies of a single template (fragment of DNA to be read). Illumina do this with bridge amplification. Ion Torrent (and others) with a process called emPCR.
In emPCR (emulsion PCR) you create a droplet in oil which is used as an isolated reaction vessel. You amplify the DNA on a solid support (bead). So in each droplet you need:
One bead
One template
Some primers
Some nucleotides
Sounds great! Except we have no way of making sure there’s exactly one bead and exactly one template in a droplet. And if you have multiple beads or templates… it gets messy.
If there’s no template… there will be no data generated.
If there’s more than one template the bead with contain amplified product from multiple templates (reads). The signal will be mixed, and again no useful data generated.
For this reason, you generally want to limit bead concentration such that there’s only at most one bead in each droplet. And just have to live with the fact that some beads will have no template and some multiple. This reduces the throughput of your platform. In Ion Torrent’s case, it seems like ~50% of wells are lost to these issues.
If you had a method that allowed 100% of beads to have product from a single template this could perhaps double throughput!
This is what Ultima appear to have been working on.
How It Works
The process is depicted in the image above (from this patent). In (1) we have a droplet containing our reagents. A bead, some templates to be read (1500 and 1502), and some primers 1504 and 1508.
We can control the number of beads in the droplet, such that most droplets have zero or one beads. We may have multiple templates. But we only want to amplify one of them.
We can also control the concentration our how two primers 1504 and 1508. Much like Illumina’s ExAmp the process takes advantage of the relative reaction rates of two different processes, in two stages.
First Stage: The original templates are extended by the 1504 primer. 1504 is present at a low concentration and this process occurs slowly.
Second Stage: Only the extension product can be extended by the primers on the beads and other primers in solution (1508). These primers are available at a much higher concentration. And extension therefore occurs at a much faster rate.
The idea is that amplification in this second stage happens at a much faster rate, and exhausts all available primers on the beads.
So, to review. The 1504 primer comes along and extends a template. Because of its low concentration this takes a long time. But it triggers rapid amplification of the template through a second primer. Another 1504 primer can then prime amplification of another template, but by the time this has a chance to happen all primers on the bead have already been exhausted.
The result is very similar to Illumina ExAmp, where a slow process (seeding of a well with a template) happens slowly and a second process (Isothermal bridge amplificaiton) happens much more quickly exhausting all the primers in a well before a second template can seed the well.
Results
From what I can tell empty well rates in the 10% range and polyclonal rates of 20 to 30% are common on Ion Torrent’s platform. This results in ~50% of wells producing usable data.
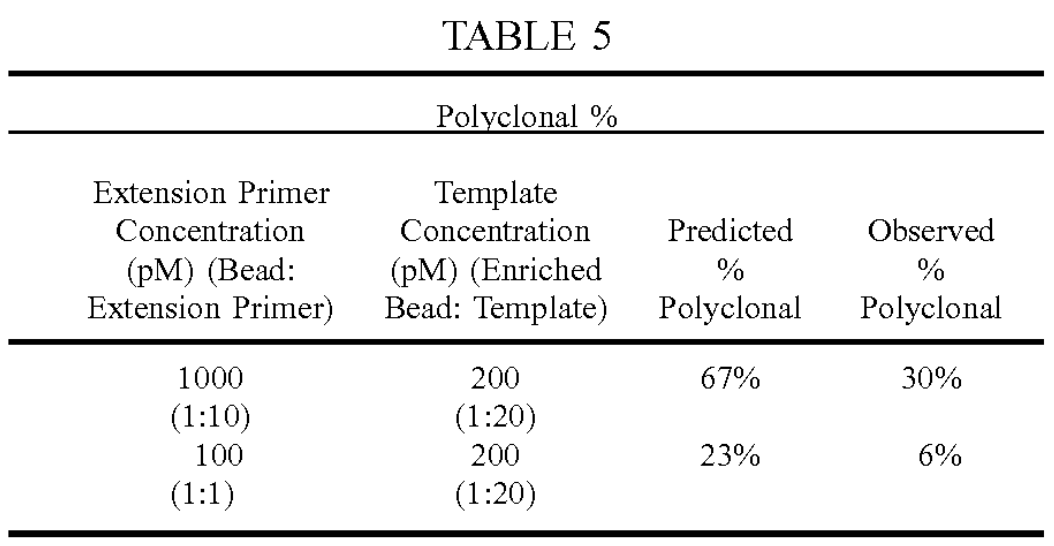
In Ultima’s patent they show polyclonal rates of <10% in some cases:
This appears to be assessed through fluorescence. You can see a nice shift from high extension primer concentration (A below) to low (B below) accomponied by a similar shift of the population away from the “Poly” group:
Summary
I don’t know if Ultima are using this in their production instruments. But they’ve clearly been thinking about ways to improve on emPCR, PPMSeq being another example where emPCR is being used to their advantage.
The approach here gives them another potential edge. Could Ultima single template rates be higher than Illumina PF percentages?
That’s less clear But I’m slowly getting sold on the idea that emPCR might actually have some benefits in spite of its bad reputation!
So while Ultima’s willingness to engage in a race to the bottom still feels like a concern, I’ll be watching with interest!