Executive Summary: There’s evidence you could do single molecule sequencing within clusters. This would be cool! And could enable things like sub-reads/long reads on clusters. Illumina clusters on patterned flowcells seem to be 10% non-read template. Which suggests room for improvement.
Illumina sequencing has this problem. It uses these little “clusters” of identical copies of the same fragment of DNA1, which is great if they are all the same fragment! But causes problems if there aren’t
On unpatterned flowcells (MiSeq, MiniSeq, NextSeq 550) clusters are randomly orientated on the flowcell. So just by chance two clusters might overlap2. When this happens the result is an ambiguous basecall. That data generally gets thrown out (non-PF).
On patterned flowcells the story is slightly different. Illumina uses a process called ExAmp (Exclusion Amplification). The surface is patterned with tiny wells3. Clusters can only grow within the wells. The idea is that if you grow clusters quickly they will fill the well before anything else can get in4.
Illumina use the term “PF clusters” to describe those clusters that are formed from a single template5 and are used in downstream analysis.
So far this is all pretty well known stuff… high PF == lots of single template clusters == good.
But this is before we discover Illumina’s secret6 single molecule metric CLUSTER DOMINATION!!!!1
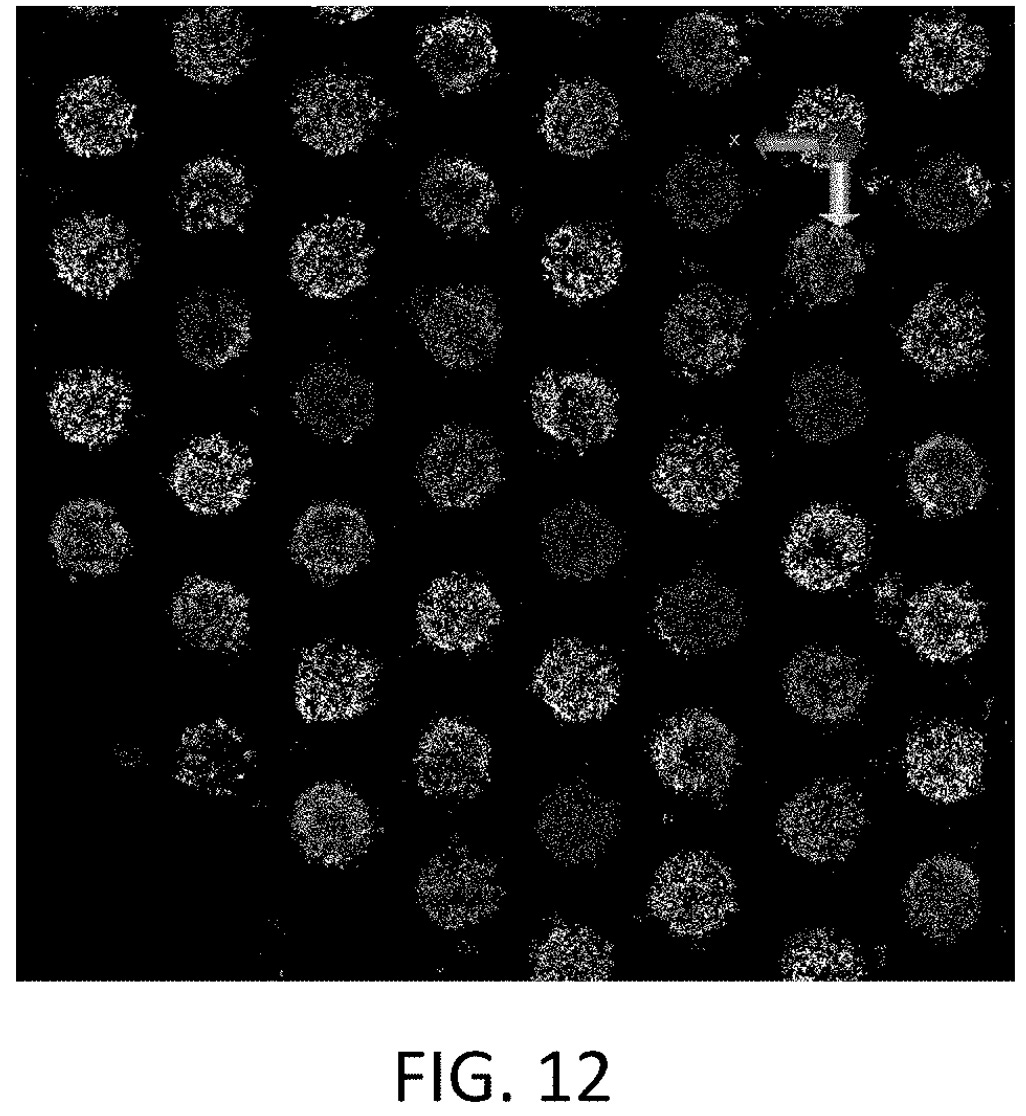
Cluster Domination
PF, original “purity filtering” isn’t really a direct measurement of the “monocolonity” of a cluster. It isn’t directly telling you the percentage of each template in a mixed cluster. It’s more a kind of “signal-to-signal” ratio. And generally a threshold is set such that you throw out clusters that result in low accuracy reads.
This was good enough for a while, and is good enough for filtering on instrument. But clearly Illumina wanted to do something better for their own development purposes.
So they performed single molecule imaging of templates within clusters. It’s kind of like they built a single molecule sequencer to sequence all the templates inside each cluster:
Well.. not exactly.