A Metagenomic Sample Prep System
In a previous post I discussed a roadmap toward ubiquitous metagenomic sequencing for diagnostics. The ultimate goal is that sequencing a sample (and detecting known and unknown pathogens) should be as simple and cheap as qPCR.
If you’re interested in the sequencing side of this take a look at the roadmap or optimized nanopore and optical approaches I’ve previously discussed.
In the roadmap I suggest that a first step would be to build out an automated sample prep solution which could be used with existing sequencers. In this post I flesh this out a bit.
The Target
Initially we would want to focus on a single target tissue/sample type. My tendency would be to look at respiratory samples via nasal swabs. I would also look at RNA rather than DNA. This will let us cover RNA viruses and bacterial infections via transcripts (this appears to work well in some contexts).
Why respiratory? Well, our goal is really to build tools to help prevent future COVID-19-like pandemics. And this seems like the best place to start.
The Prep
A single molecule approach (like Reticula) might help simplify the prep. Here we could do direct RNA sequencing and we would not require adapters if random priming was used.
However for most existing platforms we will need to perform cDNA conversion, adapter addition and amplification prior to sequencing.
I actually imagine a demonstrator being a sample prep device providing prepped Illumina libraries. This would give compatibility with a number of instruments (potentially from other vendors too).
But I’m kind of a fan of demonstrating the approach with an iSeq. This instrument keeps all reagents confined to the cartridge. This means that the whole thing can be disposed of as potentially infectious waste, and the instrument itself wiped down.
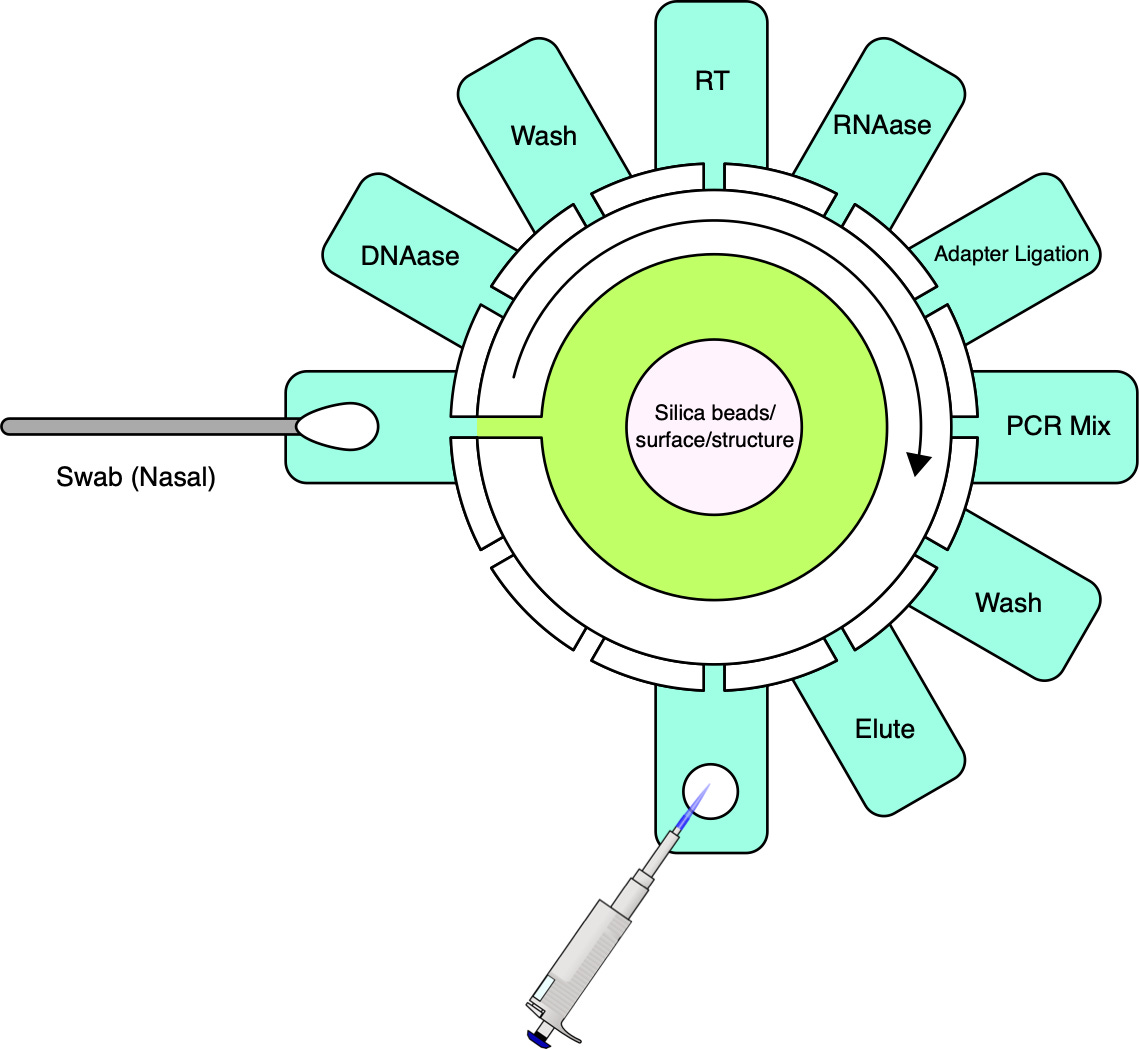
The Prep Cartridge
In a previous post, I discussed the Cepheid GeneXpert system this is a simple injection molded device which performs extraction, cDNA conversion and amplification as part of a qPCR based sample-to-answer diagnostic device. Most of these steps are common to sequencing prep too, and I therefore tend to use it as my model of a sequencing sample prep cartridge:
Cepheid GeneXpert cartridges cost about $20, though some have estimated they cost <$5 to make. I’m not fully committed to a injection molded design (alternate fluidic systems might work well). However, I’m not clear on the advantage of a digital microfludic system here (Voltrax, Voltera, NeoPrep) here. I think the increased cost of goods and limited reagent volumes are an issue here. This high cost of goods would become an issue as we attempt to get the total cost of the system competitive with qPCR (<$10).
So my default assumption is that we would want to produce a simple injection molded design which automates the required prep, using a valve and pressure (plunger) to expose the sample to different reagents:
The system and cartridge could be designed to support different workflows. But as mentioned, we would focus on RNA from nasal swabs initially. In the first instance, this might also not be a “quick” workflow. Focusing first on building a robust platform before moving from ligation to transposon or other adapter addition methods to speed up the workflow.
Other Adaptations
The device described above does not support multiplexing. The idea is to focus on a simple workflow which can be directly loaded onto a sequencer. This is also why I talk about using the iSeq, where 4M reads would be about the number required to enable diagnostics are near qPCR sensitivity for respiratory pathogens.
But we could potentially incorporate unique barcodes into the adapter addition process. These would need to be a near globally unique barcode such that any cartridge can be used with any other. To enable diagnostics we only need a read length of ~20 bp. A 15bp barcode gives us ~1B barcodes, so adding sufficiently long and distinct barcodes shouldn’t be an issue on current platforms even at the shortest available read lengths.
Summary
When I’ve discussed this above prep system with people they’ve often suggested that this could support existing work in metagenomics well (if you think this would, or would not be useful please get in touch - new@sgenomics.org). I think this is one of the most attractive aspects of this idea. It would let us engage with and accelerate existing metagenomics work.
This in turn would help us accelerate this adoption of sequencing in diagnostics and drive more interest in new platforms to better serve this market.



Interesting and educational read!
Do you think rRNA depletion could be incorporated somehow?