Sequencing: STILL More Sensitive than qPCR
Late last year I wrote a post claiming that sequencing is more sensitive than qPCR. In that post I looked a few papers I’d recently come across which seemed to show that sequencing is more sensitive than qPCR in those contexts. A reasonable concern is that I looked at a relatively limited number of papers… so, I decided to try and do something a little more systematic.
I dug through the literature I could find and pulled together 42 pathogen focused metagenomic sequencing papers, which could hopefully be used to assess the sensitivity of sequencing as compared to qPCR/Panels. Keith Robison of the excellent Omic’s Omic’s blog was kind enough to send me a few as well, for which I’m most grateful.
I then filtered though these papers using the following criteria, removing papers which had:
Less than 10 positive subjects.
Less than 10M reads average per sample or read count unstated.
Only contrived or pooled samples.
Targeted or semi-targeted studies.
I suspect the read count limit might be the most contentious criteria here. I don’t really think 10M reads is a hard requirement for Metagenomics sequencing. But in studies with fewer than 10M reads you often find individual samples with <100K. Looking through the full paper list, relaxing this to 4M reads would have added one additional pre-print study, which supports the general trend in the table below.
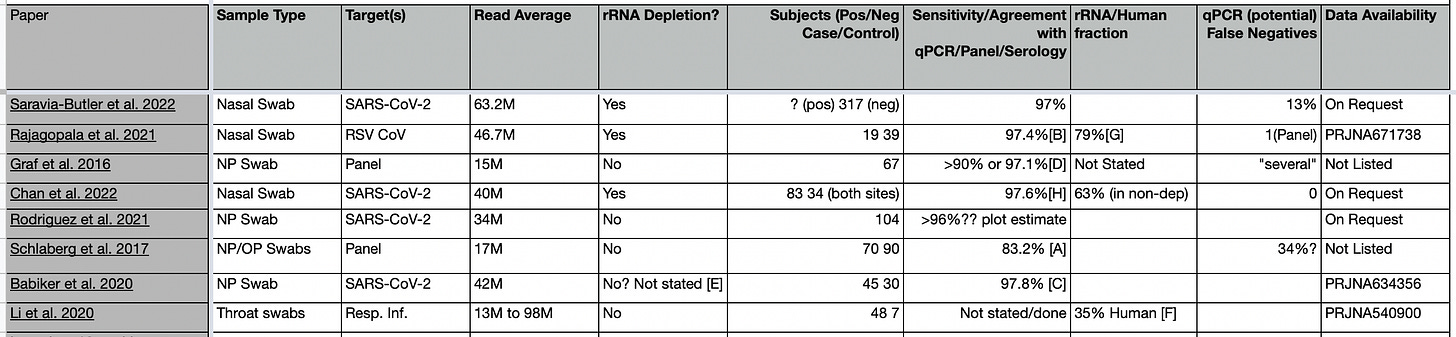
The filtering criteria left 8 papers:
You can find these in this google sheet, references A-F are comments on how these values were obtained (see the end of the post and google sheet).
Sensitivity
Three of the papers have a clear statements on sensitivity these state sensitivities (assessed via qPCR/Panels) for sequencing of 97% (Saravia-Butler et al. 2022) >90% (Graf et al. 2016) and 97.6% (Chan et al. 2022).
In three others, I calculated sensitivity/qPCR agreement from the data shown in the papers. These values were 97.4% (Rajagopala et al. 2021), 83.1% (Schlaberg et al. 2017), and 97.8% (Babiker et al. 2020).
Schlaberg et al. 2017 is the least favorable paper. They break down sensitivity by virus as follows (combining control and patient groups): IAV 0/1 (0% RNA), ADV 0/8 (0% DNA). IBV 0/1 (0% RNA), HMPV 7/12 (58.3% RNA), HPIV 5/6 (83.3% RNA), RSV 23/29 (79% RNA), HCoV 4/5 (80% RNA), HRV 46/50 (92% DNA), M. pneumoniae 7/7 (100% DNA). In total 99/119 giving an overall sensitivity of 83.1%. Not great! Evaluating these results is complicated by several factors. Firstly they were looking at both DNA and RNA targets, but only capturing DNA through transcripts. Surprisingly, both their best and worst sensitivity is on DNA targets. They also state that viruses “were detected by PCR of NP/OP swabs or serology” so this is not a clean comparison against qPCR, and it seems at least possible that we are seeing some false positives. The RSV results are particularly odd, showing 96% sensitivity in the patient group, but 0% in the control. If I had to guess, I’d say the control group used serology, and contained false positives from resolved infections… If we restrict ourselves to RNA viruses in the patient group we have an overall sensitivity of 92.1% which seems more inline with other results.
I also estimated sensitivity for Rodriguez et al. 2021, but the only data we have to go from here is a graph of Ct versus genomes copies/ng. Assuming a threshold of 1, it looks like at most 4 could be false negatives. This would give a lower bound sensitivity of 96%.
Graf et al. 2016 compared sequencing to a panel (GenMarkDx) which I believe uses a hybridization assay. Positive results agreed in 34 out of 37 samples. But then they tried to confirm the 3 potential false positives using qPCR. Unfortunately, they only had enough material for 2 of the samples, both of which came back qPCR negative. To me, this puts a lower bound of 97.1% on the concordance of sequencing and qPCR positives. Their claim is that “untargeted metagenomics has a sensitivity at least comparable to those of the RVP and qPCR”.
Personally I would tend to agree. These papers all show that metagenomic sequencing is at least as good as qPCR in my view. Where a sensitivity estimate could be calculated against PCR/Panels (5/8 papers) it was >96%. In many cases it seems likely that slight threshold tweak would be enough for qPCR and sequencing to have 100% agreement, often effecting only single samples at high Ct in relatively small studies.
qPCR False Negatives
The sensitivity of sequencing is further supported by the number of potential false negatives we see in qPCR results.
Graf et al. 2016 states that "There were several samples that were positive by both the RVP and untargeted metagenomics but negative by qPCR”.
Saravia-Butler et al. 2022 states that “42 out of 317 (13%) PCR-negative samples had detectable SARS-CoV-2 genomic material, suggesting they were false negatives”.
And Schlaberg et al. 2017 states “RNA-seq/PVG PCR detected previously missed, putative pathogens in 34% of patients.”.
I’d also refer back to my thoughts on Chan et al. 2022 where I suspect threshold tweaks might reveal qPCR false negative results.
In Rajagopala et al. 2022 they “were able to identify OC43 in a sample and recover 100% of the genome with over 23× average read coverage, which the panel did not detect”. The panel in question being an (I think end-point) PCR panel.
Overall, 4 of the 8 papers have explicit statements on qPCR false negatives.
Data availability
One of the papers didn’t provide enough information (PCR results) to estimate sensitivity or false negative rates (Li et al. 2020), but the data set is public. In total, 13 of the original 42 papers have public datasets available. A meta-analysis of these dataset seems like a worthwhile exercise! And I might take a closer look at this at some point in the future.
Conclusion
I’m afraid that this short review of Metagenomics sequencing hasn’t done much to dissuade me from believe that in the limit and many practical scenarios, sequencing is more sensitive than qPCR.
In the above studies, we’re looking at respiratory viruses. It may be the case that there are other sample types where qPCR is a clear win. However, I didn’t explicitly attempt to filter studies by samples type, they just didn’t appear in my literature search. I suspect few large, high depth metagenomics studies on other clinical samples have been performed.
But, if you know of studies I’ve missed. I’d love to take a look!
Another observation that occured to me is that indexing hopping noise introduced by multiplexing sets a lower bound on the threshold you can use for detection. This increases throughput requirements. As such platforms that can economically sequencing single samples without multiplexing are particularly desirable. What I’d really like to see is a large study where individual samples where run un-multiplexed on a low-end instrument (e.g. Miseq) this would help assess the true sensitivity limits of sequencing removing the potential for noise introduced by multiplexing.
Comments and thoughts most welcome!
Notes
[A] My calculation. Table 2 breaks down sensitivity by virus. I added together all the seq. positives and calculated the fraction of EPIC positives. The calculation is biased by the use of serology not qPCR in some cases, and the inclusion of both DNA and RNA viruses in this RNASeq study.
[B] My calculation. Paper statement "Using a very stringent threshold for virus identification (>90% genome coverage and an average read depth of 5×), we found RSV (either A or B subtype) in 38 out of 39 RSV-positive samples sequenced.
[C] My calculation. Paper statements: "Of the 30 SARS-CoV-2-negative PUIs, 8 (27%) tested positive for another respiratory virus by routine clinical testing. In all cases, mNGS identified the same virus". "SARS-CoV-2 was present in all 45 RT-PCR-positive PUIs by mNGS (Table S2). For the sample (GA-EHC-084F) with the lowest concentration of SARS-CoV-2 in our study, cycle threshold (CT) = 34, only one SARS-CoV-2 genome region was identified, which did not meet our criteria for detection by mNGS (three or more genome regions)."
[D] Paper statements: "agreement with RVP and qPCR of >90%". Paper statement "There were several samples that were positive by both the RVP and untargeted metagenomics but negative by qPCR, suggesting that untargeted metagenomics has a sensitivity at least comparable to those of the RVP and qPCR.". "In unselected samples, untargeted metagenomics had excellent agreement with the RVP (93%)." Untargeted metagenomics detected 86% of known respiratory virus infections, and additional PCR testing confirmed RVP results for only 2 (33%). Of the 37 swabs for which respiratory virus was detected, 34 (91.9%) also had virus detected by untargeted metagenomics (Fig. 1D). For two of the remaining three samples, there was sufficient sample to attempt qPCR confirmation of RVP results. In both cases, qPCR results agreed with those of untargeted metagenomics.
My reading of the above is that for the untargeted dataset. 37 Swab were positive via the panel. Sequencing agreed with all but 3 of these. Giving two potential false positives. But when they performed qPCR, they didn’t come out positive anyway. Suggesting that the the panel result was a false positive. There was one additional sample which they couldn’t perform qPCR on because they didn’t have enough material. If we use qPCR as the “standard” then we come out with a sensitivity of 97.1% for sequencing here.
[E] Paper references two citations stating "Samples underwent RNA mNGS as previously described". These papers reference two other publication, one of which contains multiple methods which includes polyA and rRNA depletion.
[F] I extracted data from the pdf table in the sup. info. spreadheet here: https://docs.google.com/spreadsheets/d/16tYMBCu3_I5z7rl_3PO-hu5fe6MzlSJKX-BWTjry-eo/edit?usp=sharing
[G] My calculation from the statement "There was an average of 46.7 million paired-end reads and 1.3 million single-end reads per sample after quality trimming. After removal of human rRNA, mitochondrial RNA, and bacterial rRNA, reads were partitioned into two bins: a human transcript bin with an average of 36.9 million reads"
[H] Combining sites A and B. Single site sensitivity was 98.4% and 95%. Both sites had a single false negative from sequencing. In the case of site A this appears to have been one of two technical replicates.

At extremely low concentrations, a perpetual concern in genomics lab is cross-contamination between samples. Hopefully labs that are running lots of qPCR set things up to prevent those sorts of situations, but could the sequence data itself help eliminate cross-contamination? For samples that are negative by qPCR but positive by seq, does the seq data support a lineage different from other samples in the same batch?
I hope to find some time to dive into these papers to see if they have addressed this...