Reticula Pt. 3 - Technological Approach
In the first post on Reticula I described my fund raising adventures and in part 2 I described the motivation for low cost ($1 COGS, $500 instrument) sequencing.
UPDATE: I’ve raised a small amount of Angel funding for Reticula, so things are moving forward. If you might be interested in hearing more please email me (new@reticula.bio).
In this post I’ll layout the overall technological approach. First, I want to take a few things off the table. ONTs 2021 accounts state that “gross margin percentage fell to 41.2% (2019: 49%)”. Their cheapest runs would be on the Flongle at $90 per flowcell. We can estimate a ~$50 COGS here. It’s reasonable to say that we can’t get a $1 COGS run here, making ONT uncompetitive against qPCR (~$1 COGS).
Similarly Illumina’s cheapest runs are >$500 assuming these are something like 90% profit, we’re still looking at a $50 COGS.
There are a few things that push up this COGS for Illumina. Firstly, there’s a fairly complex reagent cartridge. Plastics and the reagents themselves likely cost very little. But with a 14 component reagent cartridge, it’s difficult for me to see how you can further optimize this to get it below $50:
With the Illumina approach you need reagents for cluster generation (bridge amplification), cleavage reagents, second read reagents etc.. none of which to can easily do without.
For Reticula we want to remove all this complexity. In fact, we want to remove the fluidic system completely.
Not only would this simplify the consumable, but result in a simpler, smaller instrument which doesn’t require field support. If it breaks, you just pop it in a jiffy bag and send it back (much like an iPhone).
Sequencing Approach
The approach we take is single molecule sequencing-by-synthesis. Rather than cleaving dyes, we remove them with photobleaching. There’s prior work where photobleaching has been used to remove signal for single molecule sequencing. But the Reticula approach is slightly different. In prior work, photo-bleaching was taken to completion before flowing in new nucleotides.
With Reticula, we just allow nucleotides to incorporate in real time. This limits us to a single observation area, but means we don’t need to use a cyclic chemistry/a fluidics system:
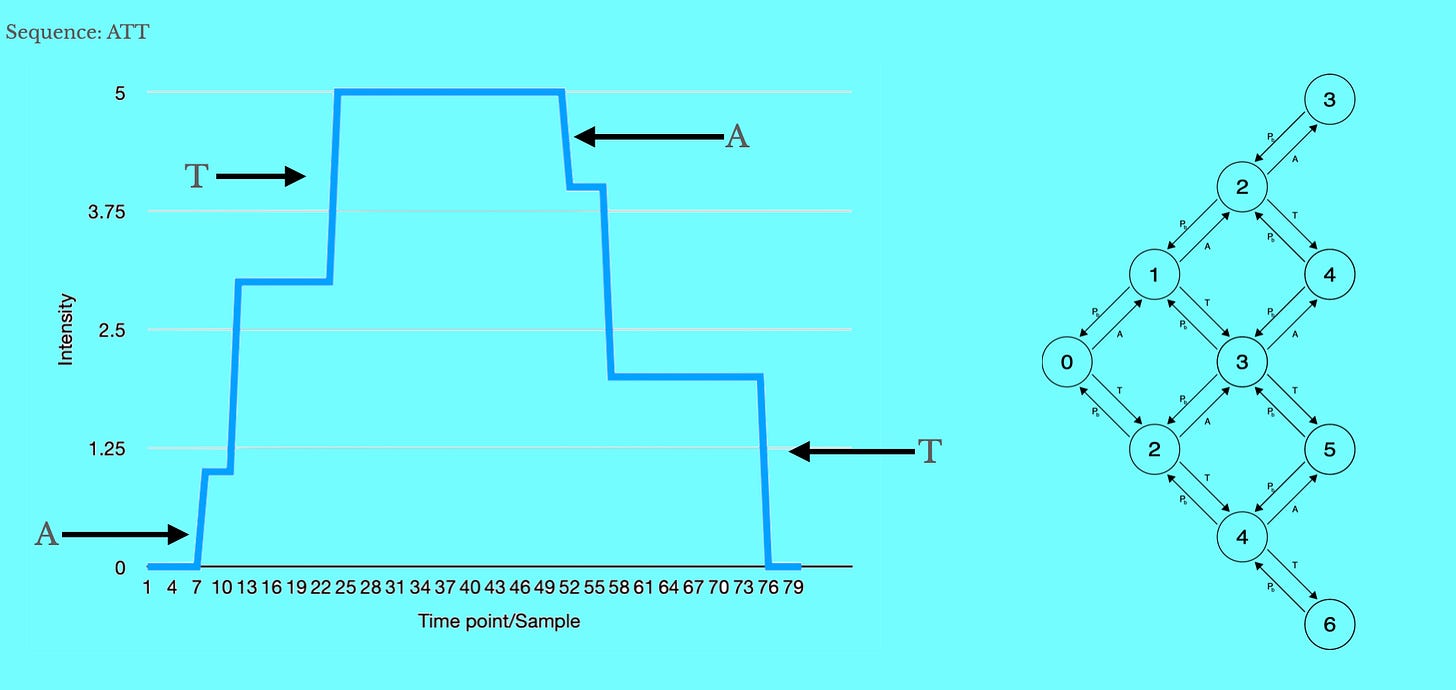
The result will be a sequencing approach where we see stepwise increases in emission intensity when nucleotides are incorporated, and stepwise decreases when bleaching occurs.
To determine which nucleotide was incorporated we can use one of two approaches. Either each nucleotide is labeled with a dye that emits at a different wavelength (the traditional approach) or we select dyes such that they different in emission strength at a single wavelength.
The advantage of the second approach is that we only need a single filter/camera, further simplifying the instrument.
The result will be a somewhat convolved “steppy” signal. Upward steps are incorporation events, downward steps are bleaching events. Bleaching events can occur in any order, but incorporation events will obviously occur in template order.
However, both incorporation and bleaching events give us information about the template being sequenced. An HMM can be used to combine all this information and find the most probable underlying template sequence:
The system described above doesn’t need any fluidics. You can imagine for example that your polymerase and nucleotides might be directly embedded in the flowcell. When the sample is added, it could wick into an imaging area and the polymerase would start incorporating bases. A fixed camera would then be used to observe incorporations in real time.
Optical System
To make this work we need a cheap platform for performing single molecule experiments. Back when Helicos was around, this wasn’t really possible. And even today most research users use SCMOS cameras which cost $20000++.
But it turns out consumer grade cameras have got a lot better:
For my experiments I’ve been using IMX178 and IMX174 cameras. These cameras are designed for low light security applications. They work well and cost <$300 in single units.
To observe single molecule events, we really need some kind of TIRF setup. Most current research experiments use objective style TIRF. The problem here is that high NA TIRF objectives are expensive ($5000). So we don’t want to do that. Instead we use prism style TIRF.
Rather handily this was the approach used in the original Solexa Genome Analyzer. So my prototypes repurpose this optical system:
This setup has allowed me to prove out the optical approach and obtain single molecule photobleaching data:
I’ve also sourced cheap fused-silca prisms, lasers and objectives from Shenzhen. Overall, this suggests that it should be possible to build a very cheap, small instrument:
The one issue with prism TIRF (as anyone who has used a Genome Analyzer will know) is that it’s a bit messy. This is because you need do put oil on the prism to couple it to the flowcell. You may be able to get round this by using a plastic prism embedded into the flowcell, the issue here is that most plastics are somewhat autofluorescent. Some plastics however, are better than others. And you maybe able to find one where this isn’t an issue.
Summary
In this post I’ve discussed the Reticula approach, and why I think it should result is a very low cost of goods of ~$1 per run. The approach would likely be limited to 1M reads over the fixed imaging region. You could potentially push this further, for example once everything is bleached out, you could imagine adding more sequencing templates.
Similarly, photodamage to templates is going to limit read length. Other (Helicos-like) sequencing platforms have reached reads in the 33 to 53bp range. We’re also going to have comparatively high error rates, without further nucleotide development probably in the <5% range.
It’s possible that those specs can be improved, but as discussed in the previous post, they are more than good enough for viral diagnostics. Hopefully the compelling thing about the Reticula idea, is that above all else, we try and keep the cost per run as low as possible.







Hi Nava, this is a very interesting minimalist idea.
Some thoughts: I assume you'll try to keep the nucleotide concentration very low so that simultaneous incorporation of multiple bases is minimized (the first being bleached well before the second is incorporated), as well as the background fluorescence from bulk nucleotides?
Assuming you're capturing the templates on a random n-mer oligo lawn, is there a possibility that more than one n-mer will bind and prime the same template, be simultaneously extended, creating mixed signals?