Many thanks to those who provided suggestions and assistance after my last post some interesting options appear to be available and all suggestions and comments are much appreciated.
Today I’m going to talk about an old MiSeq I bought on eBay, and my experience getting a PhiX validation run completed.
The instrument was sold as “for parts” and had no login information. It was therefore a bit of a gamble… MiSeq’s still sell for >$100K new and >$10K on eBay. But being “for parts only” this one was significantly cheaper.
Paid Subscribers help justify the cost of this purchase and I’m grateful for their support.
Before getting the instrument running there were two major issues that needed to be resolved:
Getting Illumina to let me buy reagents.
Getting the instrument shipped to Japan.
I think Illumina would rather I buy a new iSeq, but this would have been a more expensive initial purchase in addition to the kits costing significantly more. A new MiSeq would be impossible… but they did agree to sell me reagents after some back and forth.
Having never organized freight shipping, figuring out all the customs issues was painful… but we got there in the end.
Password Reset
MiSeqs are heavier than they look… but after a certain amount of struggling I did manage to get it up two flights of stairs… the next problem was logging into it… none of the standard passwords worked so a password reset was required.
This turned out to be pretty easy. The MiSeq used a single 1Tb SSD. I mounted this drive on a Linux system and used chntpw to reset the passwords. From memory you also need to change configuration for one of the services (Universal Copy Service?) to use the new password you set.
With done the instrument was at least theoretically functional.
Washes
Luckily the MiSeq came with wash tray and an old PR2 bottle. Both had a thick layer of sticky looking Tween residue. I washed these pretty throughly with lab grade water1.
Next I prepared Tween solutions as described in the MiSeq System User Guide2 and ran the maintenance wash.
PhiX Control Preparation
I bought some PhiX control from Illumina in order to perform a validation run3. I prepared this as described in the MiSeq “Denature and Dilute Libraries Guide”. This requires NaOH and Tris-Cl. Thing is I didn’t have any Tris-Cl only Tris-EDTA. EDTA is supposed to cause issues for polymerases. However as it’s going to get washed through the system anyway I decided it was probably fine… (and I’d already thawed out the sequencing kit).
For reference here are the reagents I used:
With the PhiX prepped I loaded 600uL of it onto the cartridge which had been thawing over night in the fridge.
Sequencing
The cheapest MiSeq kit appears to be a 300 cycle v2 nano kit. I figured it would be fun to run a full 300 cycles and perhaps “try some things out”.
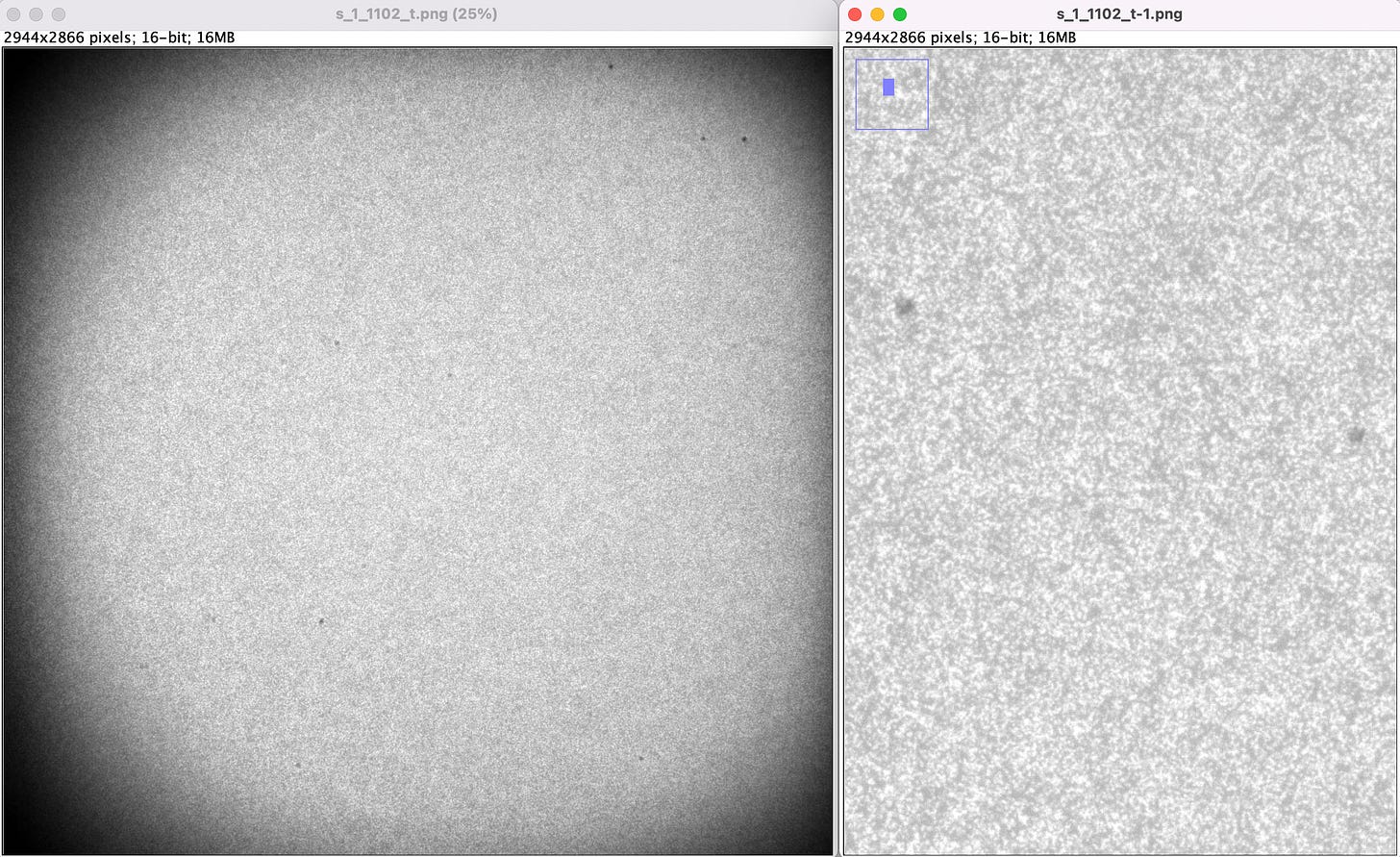
Well… I guess shouldn’t have worried about EDTA stopping clusters from growing as the run was pretty over-clustered (at least that’s how the images look to me):
I didn’t end up running the full 300 cycles (for various reasons… ping me if you want to discuss weird MiSeq stuff). But I did get a solid 20+ bases off which aligned to PhiX reasonably well.
So I guess the MiSeq works and I can now add sequencing to the consulting services I offer. :)
I have a few more projects planned for the MiSeq which paid subscribers might help fund. If you have any specific thoughts on interesting and tractable projects ping me on the Discord4!
Well actually I just went down to the local chemists and bought “high purity water”… but it seems fine.
Basically 500ml of 0.5% Tween. Again I used “lab grade water” for this as described above. I’m using whatever lab ware I have knocking around at the moment. Here this includes some 50ml centrifuge tubes I’d bought for some unknown purpose on eBay many years ago and some 12ml syringes as I don’t have a decent pipette in the ml range (consider a paid subscription to support the use of proper lab equipment).
Down to 12.5 pM for a V2 kit.
Or by email: new@sgenomics.org if you prefer.






I think we've come full circle now, as the first Solexa 1G's in 2007 had 26 bp readlengths (that is if memory serves correctly). Will never forget my first customer at NHLBI, what an effort it was to keep that instrument running.
Solexa had as part of its 'acceptance criteria' 1 Billion mappable bases (thus the name "1G") but those first instruments were lucky to get 800 Million bases (800 MB) of mappable reads.
It is notable that no one ever returned one!
Nice recycling. I hope others are inspired by your efforts to bring MiSeqs to labs who wouldn't otherwise have the budget. Of course, the budget to run sequencing is another factor... it comes back to an old adage, "there is no such thing as a free kitten".