Over on the Discord someone posted this image (originally from LinkedIn):
With the commentary of “what diagnoatic yield would be significant enough to quieten the haters?” and an ensuing discussion which contained “more heat than light”.
I was asked for my input. So, I’m going to write a few thoughts here, but in summary:
I’m strongly in favor of long read sequencing as a first diagnostic genetic test in rare disease testing. The price differential is minimal the increase in diagnostic yield is clear. I also can’t see that short reads are “better” than long reads1 in any way that counts.
I also think that driving adoption will be extremely hard. A suspect only a tiny fraction of rare disease patients have any genetic testing. My guess is that clinicians will continue to perform karyotypes, arrays, do exomes, then short read whole genomes, and finally maybe maybe long read genomes.
This sucks, and is in part due to institutional resistance to testing when it doesn’t result in a change in patient care2.
What’s Missing
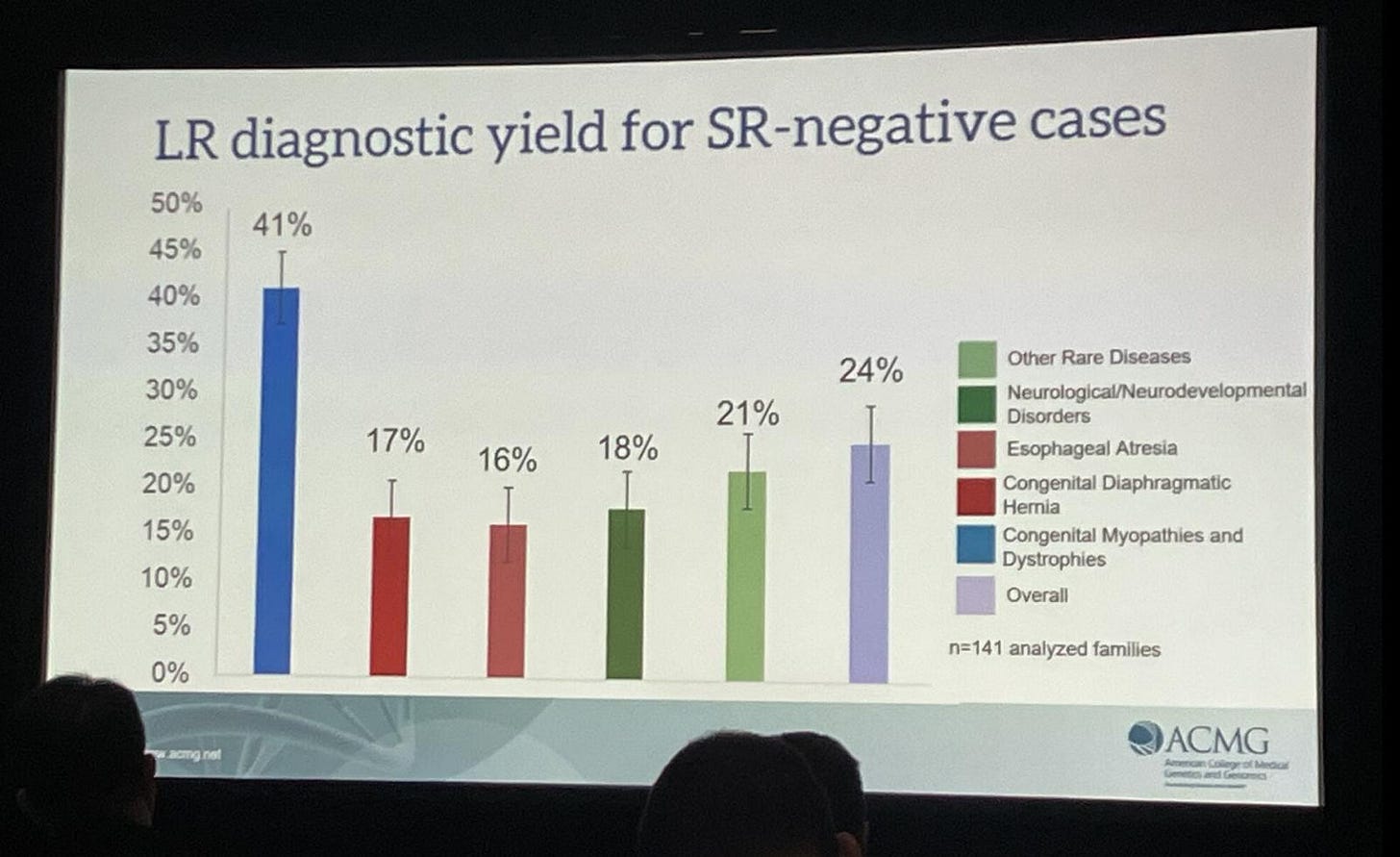
There are a few things that make it difficult to draw strong conclusions from the single slide above. The first is, we don’t know the total cohort size and yield from short read sequencing. One study suggest this should be about 41%. This being the case, you’d go from an overall diagnostic yield of 41% to 55% by moving to long read sequencing.
The other issue is that the slide above doesn’t tell us how many of the new variants could only have been identified with long read sequencing. Reanalysis of whole genome sequence data often results in an increased diagnostic yield of up to 34%.
I’ve seen some of the other slides from the presentation above. It appears of the total 24% increase, about 10% could only have been identified via long read sequencing. I assume the rest is therefore due to reanalysis including more pathogenic variants/better methods.
So, we have some unanswered questions, and the yield increase could be a little lower than the headline number…
The thing is, these results are far from unique and increased diagnostic yield from long read sequencing has been demonstrated by a number of other studies…
They’re Not Alone
Results showing increased diagnostic yield from long read sequencing are not uncommon. This study showed a diagnostic yield of ~10% for long reads in cases which were short read negative. Another review showed 7 to 17%.
In neurodevelopmental disorders long read sequencing was used in 2021 to identify possible genetic causes previously missed with short read sequencing. The same group published a paper in 2024 with a group of 96 patients, 7 patients showed likely pathogenic variants which could not be identified by short read sequencing. An ~7% increase in diagnostic yield.
Overall a 10% increase in diagnostic yield using long reads seems reasonably consistent with the literature3.
Short Read Is Cheaper
One argument might be that short read is cheaper. My estimate of GeneDx costs for short read.WES/WGS was >$2500. Others have suggested $4k to $7k. These numbers suggest that sequencing is a fraction of the overall cost.
But ok… let’s assume we’re talking about an additional $1000 for long read sequencing. Running the number we find that you’d be spending less than $100M to go directly to long read sequencing for all cases in the US4. If I use my conservative $2500 short read and $3500 long read costs I get:
You can triple these numbers for Trios. And double them again if you think my numbers are too conservative. But whatever you do, costs are going to be in the mid-100Ms to low billions.
This seems like a tiny amount of money in comparison to the other costs associated with these diseases (~$1 trillion a year). In a healthcare system that spends $4.9 trillion annually.
So whether you’re take make a humanitarian or purely economic argument. Aren’t these costs worth it to better serve not only these patients, but to potentially accelerate treatment development and improve the lives of future generations?
I don’t get the cost argument…
But…
If you break down Illumina’s clinical markets, only about $500M in total is going to Genetic Disease Testing. Companies like GeneDx have revenues in $300M range, but only about 34% of their test volume is WES/WGS. They appear to have run ~70000 WES/WGS tests in 2024. I suspect GeneDx are a market leader here, and yet this number is only a small fraction of the estimated new rare diseases cases. And of course, they are using short read sequencing.
So it seems that we’re a long way from seeing universal adoption of short read sequencing in rare disease.
I’m hopeful that we will one day see clinical long read sequencing routinely used in rare disease diagnostics. But I suspect we’re looking at time scales measured in decades rather than years.
And it may well be the case that many clinicians will hold off on adopting these approaches until as new therapeutic options become available… which convince them that these tests not only offer valuable insights, but result in improved patient outcomes.
The benefit of PacBio long reads over short reads seems like a clear win. For ONT it’s less clear due to overall error and systematic bias. So I wouldn’t personally be able to make this call as easily.
This is somewhat ridiculous in my view, as at the most basic level identifying a genetic cause may affect family planning and siblings.
Please ping me (new@sgenomics.org) if you have more/better numbers.
I’m going to estimate cases based on births. Currently there’s probably a huge backlog of un-sequenced individuals. but perhaps more interesting to understand the potential ongoing costs, than the backlog. Rare diseases affect ~5% of people. There are ~3.5M births a year in the US. Giving us 175,000 cases/yr. So, if a long read test costs $3500 and a short read $2500. Then:
All Short Read Only: (175000*2500)/1000000 = 437M USD.
All Long Read Only: (175000*3500)/1000000 = 612.5M USD.
Short then Long: (((175000*0.41)*2500)+(((175000*(1-0.41)*3500))))/1000000 = 540.75M USD.




Radboud University in Netherlands - which as I understand it essentially handles all rare disease testing in that country - has gone to PacBio WGS as first line testing since it eliminates the need for nearly all of the other technologies required to complement short reads and with it the training complexity & upkeep costs of maintaining competency in all those techs
http://omicsomics.blogspot.com/2024/05/hifi-wgs-as-nearly-unified-tool-for.html
https://pubmed.ncbi.nlm.nih.gov/39809270/
https://www.medrxiv.org/content/10.1101/2024.09.17.24313798v1
HiFi still struggles with a handful of mutation types - Robertsonian fusions, regions with long purine tracts on one strand - but that means retaining just a few complementary technologies.
On the cost side, false positives / ambiguous calls from short reads must also be considered & the expensive personnel time to review these
I’d agree that institutional conservatism is the biggest barrier to long read adoption
I'm surprised that short read tests cost $2500, these days I think the actual sequencing costs are much cheaper (probably at most $800 for 30X coverage, including library prep, and I've seen cheaper estimates than this). So is the rest just profit margin for the provider?