Pleno Inc. - PCRquencing
About a year ago I engaged in some pretty baseless speculation on Pleno. Since then they’ve raised $40M more, bringing their total to $55M. But more interestingly, a couple of patents have been published!
The best I can claim for my previous post is that they do appear to be “incorporating multiple pieces of information from each probe”. But via an approach that is very much like sequencing. Let’s take a look at these patents and see if we can figure out what’s going on!
Fluidics, cartridges and Sample Prep
Over on GenomeWeb, Pleno made some explicit statements about their approach:
Pleno's platform is based on a set of reagents it calls plenoids — DNA probes to specific targets that are each linked to unique DNA-based codes called Hypercodes. Users add these probes to their sample (in a well of a 384 or 96 well plate) and after the probes hybridize to their targets they amplify the specific Hypercode linked to each probe. They then label these Hypercodes with conventional fluorophores and optically read out the specific signal generated by each of these Hypercodes. Amplification is done using a technique that Gavin Stone, Pleno's VP of product development, declined to disclose but which he said was a non-PCR nucleic acid amplification approach currently in use in the broader research community.
That amplification approach seems to be rolling circle amplification1. This allows them to create little nanoballs of amplified DNA labeled with a “code” (more about that later). By “broader research” community I assume they mean that this approach has been widely deployed on MGIs sequencing platform.
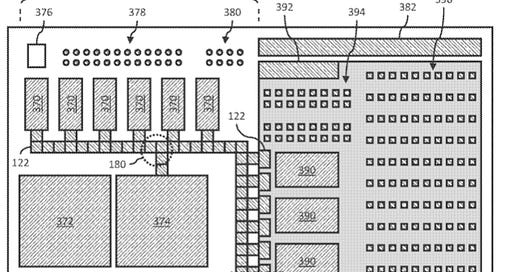
Pleno also describe a fluidics cartridge used to integrate the sample prep and detection process. This seems to be a pretty traditional digital microfluidcs (DMF) approach, similar to Volta, the NeoPrep, Voltrax etc.
Their system seems like it uses a hybrid PCB and CMOS chip based DMF solution. PCBs would be cheaper, but a CMOS based DMF potion would let them use smaller feature sizes and potentially integrate detection into the device.
From the GenomeWeb statements we know this is an optical fluorescence readout.2
So, we have these padlock probe based nanoballs (a seemingly existing approach) with a short “code” on them. This is a short, known DNA sequence.
Now all we need to do is read out those sequences. What could we call that reading process? Sequen… NO! HYPER-DECODING! We’ll call it hyper-decoding!
Decoding
The patents specifically mention using pyrosequencing, and optimizing codes for readout using pyrosequencing. Codes can be designed such that homopolymers3 do not encode information. In addition to this, you can design your codes such that are a good match for the flow order being used during sequencing. Your sequencing flow order code be anything e.g. ATGCTGAC. The code ATGCTGAC4 would read out as 1111111. TGCTGAC would read out as 01111111. CA as 00010010 etc.
The point is that it’s not the length of the code that matters, but how many cycles are required to decode it.
If you have control over both the encoding and the sequencing approach, it makes sense to optimize them for efficient encoding/decoding.
While the patent mentions pyrosequencing the general approach would also apply to unterminated ISFET or single channel optical approaches.
In order to demonstrate this process, Pleno use a traditional sequencer (looks like a MiSeq to me):
Summary
Pleno’s idea is to compete with PCR where PCR doesn’t scale to enough targets. This kind of steps them up against the FilmArray and similar platforms. Of course the patents mention multiple approaches, and they could also be using targeted arrays. But the public discourse around “hyper coding” suggests that they are pursuing the “sequencing-like” readout discussed above.
This presumably means that their CMOS device and fluids system incorporates the necessary reagents to perform a limited amount of sequencing, say 16 cycles5.
It’s a fun idea! A multiplex PCR-like system with a mini-sequencer for readout!
After the paywall break, I discuss some of the challenges here, and how the approach fits into the broader market.
Just Build a Sequencer
Personally I think if you’re going to build a sequencer… just build a sequencer.
Technically, I don’t see anything impossible about the Pleno approach much of it has been demonstrated in other contexts. But I’m not so clear as to why you would do this…
In particular… I can’t see any advantage to this approach over just integrating the necessary sample prep approach into an iSeq. The DMF system and CMOS sensor used here is unlikely to have a lower cost of goods than an iSeq cartridge. You could perform 8 cycles of sequencing on the iSeq and get the same code based readout… so why build a funky limited sequencer, when you could build one that could address broader applications?
This perhaps reflects my personal biases around sequencing being a universal readout…
As Sequencer…
As a sequencing device, the instrument looks somewhat similar to the Genapsys system, with a disposable CMOS chip. As a standalone system this probably doesn’t have a great product-market fit. Genapsys clearly failed here, and other “cheap sequencers” have not been wildly successful.
I think the market for a cheap sequencer where consumables cost ~$100 is just too small. Pleno, with a relatively complex flow cell would I suspect still be relatively high cost.
Plano’s sample-to-answer components of the Pleno could help here. A cartridge providing a sequencing based readout, costing a few hundred dollars might find a niche….
Multiplex-PCR Markets
Looking at Pleno from a market perspective it would presumably be competing with higher “plex” PCR platforms like the FilmArray. The Pleno approach looks significantly more complex and I’m not confident that they could approach the FilmArray COGS (<$50?) using a DMF system and disposable CMOS sensor.
If you’re going to go the “custom sequencer readout” route, why choose this one? At relatively low throughput wouldn’t a glass flow cell and fluorescence readout work equally well?
This then starts to sound an awful lot like a custom Miseq… which begs the question… if you are successful, couldn’t Illumina (or another sequencing company) build something very similar potentially with a lower COGS.
Overall… I think the Pleno approach is fun! But I don’t see how it fits into the market…
Or optionally bridge amplification, or a bead based amplification.
Runs of the same base.
Would be the complementary on the template being sequenced, but you get the idea.
This would give headroom for 10,000 targets+error correction, 65536 possible states. Each “cycle” is either “on” or “off” and codes are designed such that they switch cycles “on” or “off” so we’re not looking at a base 4 system here really.