I’ve been pondering the implications of Roche’s nanopore announcements on Oxford Nanopore. There are essentially three ways this effects Oxford Nanopore (ONT) that I can see:
Now: How does this compete with Oxford Nanopore and as it stands today?
Future: Does the future potential of Roche’s platform compete with Oxford Nanopore?
Potential: What does the technology Roche has developed tell us about the potential for nanopore sequencing in general?
Now
As it stands Roche’s platform generates relatively short reads, is not capable of direct RNA, and is not a portable instrument (like the MinION1). As such it isn’t really directly competitive.
Roche could play in fast(er) time to answer applications. Even with a relatively long sample prep process, it’s still faster than Illumina (but likely slower than ONT, at least in some scenarios).
Beyond this, Roche’s approach likely has a lower COGS/Gb than Illumina. If ONT were trying to get near cost competitive with Illumina to open up more applications, they now have an even lower cost to aim for.
Future
Roche’s nanopore platform generates short reads, can they generate long reads in the future?
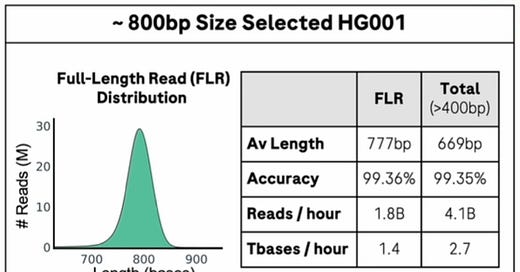
The Roche presentation showed runs with 800bp average read length and an individual read of 2284bp.
I’d guess that the current system can probably generate 1 to 2Kb reads at some reasonable throughput with a Q20 accuracy.
Midi-reads maybe become a thing but I’m not convinced by their marginal increased utility over regular long reads. If there was more value here, Illumina would be more regularly pushing toward 1Kb reads on their sequencers2.
What we don’t have a clear picture of is where the Roche sequencers peak if given un-size selected templates. Or how far they might be able to push this.
We’re therefore left to speculate… Illumina sequencers launched with 25bp reads, we’re now at 250bp+. A 10x improvement over launch isn’t unprecedented.
Personally I think it’s somewhat likely that in the medium term we’ll see Roche sequencers capable of producing a reasonable population of ~20Kb reads.
This would become a competitive problem for both PacBio and ONT. At this length, I’d expect to be covering the majority of clinically relevant applications requiring long reads.
ONT will still have utility in direct RNA (which I can’t see that Roche can address with their current approach) and very long read research apps (50Kb+).
Potential - Density
It’s what Roche tells us about the potential of Oxford Nanopores approach.
Can we run Oxford Nanopore style strand sequencing on Roche’s chip and get super-long reads at high throughput?
Probably not.
There are two advances that Roche has shown over ONT on the chip. The first is a combined ASIC and nanopore array. Oxford use a 3 layer system combining a nanopore array with TSV vias PCB and ASIC:
Roche have replaced this with a single ASIC with integrated wells. This has two advantages. Firstly in ONTs system density is limited by practical considerations around bonding chips to PCBs. ~100 microns is as dense as you can get and will therefore always limit the density of Oxfords chips.
Secondly the fabrication costs for a single ASIC/nanopore array should be significantly lower.
The Roche approach shows that it’s practical to do everything on one chip. Potentially reducing COGS by ~50%.
Next let’s look at density. Roche use ~5 micron wells, Oxford 150 micron. In part that’s due to the fabrication process mentioned above. But another issue is that well lifetime scales with well size for Oxford:
Based on the image above from ONT patents, you’d be looking at a well lifetime measured in minutes for a 5 micron well. At this lifetime, ONTs chips would be generating essentially no useful data.
Roche are showing 7+ hours of sequencing at 30 times ONTs density. How?
In part I think this comes down to the Roche’s use of Expandomers. The Expandomer process separates over the voltage used to drive the strand through the pore and the voltage used for sensing.
It’s continuous DC current that appears to deplete the electrochemistry and determine the lifetime of the well. Roche avoid this by only using brief voltage spikes to advance strands through the pore. Movement is otherwise blocked by a “translocation control element”.
During the detection period an AC voltage (thanks to Discord comments for helping me understand this) can be applied to measure the current blockage. Because that doesn’t drive the ionic current in or out of the pore, it doesn’t deplete the electrochemistry3.
Oxford just can’t do this…
Potential - Noise
Putting aside the density/pore lifetime issue. Does the Roche chip show other attributes that would make it practical or useful for Oxford-style strand sequencing?
Not really.
The traces shown suggest a sample rate of ~1.8KSPS. The MinION appears to capable of sampling up to 33KSPS. More generally they run at 4 or 5KSPS. While the Roche chip may be able to run faster than this4. They don’t really need to.
They can also tolerate much higher noise levels than Oxford. The Roche system only reads out 4-levels. Based on the traces they’ve shown, I’d estimate these cover ~200pA5. Approximately 50pA spacing between levels.
In Oxford’s system you’re trying to cram many more states (1024) into a smaller range (probably 40pA). A much harder problem.
Roche can therefore tolerate much more noise, and it’s seems likely that they have engineered a system with a higher baseline time, perhaps optimizing for more space efficient amplification circuitry6.
Summary
The Roche chip specifications and expansion approach are tightly coupled. This makes it difficult to see how they could be applied to Oxford-style stranded sequencing in the short term.
Longer term, perhaps it’s possible for others to also develop approaches which decouple translocation and sensing7. But given how long it took for Roche to get their platform up and running this doesn’t seem likely in the short term.
But with Roche’s demonstration, it feels like we have reached a tipping point in single molecule sequencing. There may be many more exciting developments to come from Roche and others.
Oxford financials released yesterday show that MinION sales have been declining. So perhaps this isn’t a very compelling market anyway.
Seeing as they demonstrated 650bp+ reads way back in 2012, this seems totally possible.
This is described in Roche patents for example:
“However, operating a nanopore sensor for long periods of time using a direct current can change the composition of the electrode, unbalance the ion concentrations across the nanopore, and have other undesirable effects that can affect the lifetime of the nanopore cell. Applying an alternating current (AC) waveform can reduce the electro-migration to avoid these undesirable effects and have certain advantages as described below. The nucleic acid sequencing methods described herein that utilize tagged nucleotides are fully compatible with applied AC voltages, and therefore an AC waveform can be used to achieve these advantages.”
we don’t know.
This could rely on different pores, or motion control enzymes perhaps rather than an expandomer-like translocation control elements on the strand.







Illumina’s roadmap is now talking about 2x500 reads
But they probably won’t go much further, as 500bp is about the limit for the current patterned flowcells (my wet lab colleagues cued me in to this) - don’t know if that is well geometry or the enzymology, but inserts over that size are disfavored
What you described from Roche is 15 years of work, without having to commercialise, it is still materially behind ONT and at best competitive with ILMN or Ultima