Other Nanopores - Part 1
A few posts back I discussed Ionic current nanopores, including biological nanopores as marked by Oxford Nanopore and Qitan and the fabrication processes used in these platforms.
In this post I want to review “other” approaches to nanopore DNA sequencing. It should be noted that none of these approaches have shown anything like the success that ionic current nanopore sensing has and have yet to demonstrate even proof-of-concept on sequencing.
In particular in this short review I’ll be focusing on startups investigating tunneling current, conduction, field/charge and optical nanopore approaches (if I’ve missed anything please reach out!).
Tunneling Current
Several companies have worked on tunneling current approaches to DNA sequencing in particular: Quantum Biosystems (where I previously worked and which appears to be dead) and Genvida.
Tunneling is ability of electrons to “jump” across a gap where conventional conduction would otherwise not occur. The technique is used in scanning tunneling microscopy (STM) to image conductive materials. Here a conductive tip is scanned within nanometers of a surface to build up an image.
While a number of publications show DNA imaged in a conductive surface, there’s much less support for the detection and classification of individual nucleotides when DNA is imaged.

In any case, sequencing using tunneling currents has been pursued by a number of groups. This mostly takes the form of embedding electrodes in a nanopore or building a nano gap structure. Here the DNA translocates through the pore, an interferes with the tunneling current:
Academic work has shown large increases in tunneling current as DNA translocates through the pore. However as previous work, and simulations suggest that it’s difficult to detect individual nucleotides using bare electrodes. Genvida have therefore been looking at functionalizing electrodes in a number of ways.
Current differences between nucleotides are likely to be in the pico amp range. As such, you will need to slow the translocation of the DNA in order to detect these small current changes. This in itself may prove problematic. In particular, using an enzymatic motion control strategy might interfere with the tunneling current, adding noise. And we’ve yet to see a demonstration of enzymatic motion control on a solid-state platform in any case.
Back bone Conduction
I’ve previously talked about Roswell Biotechnologies. Due to their use of a nano gap across which they sense currents, I’d previously lumped them in with tunneling current based approaches, but this may not be entirely appropriate.
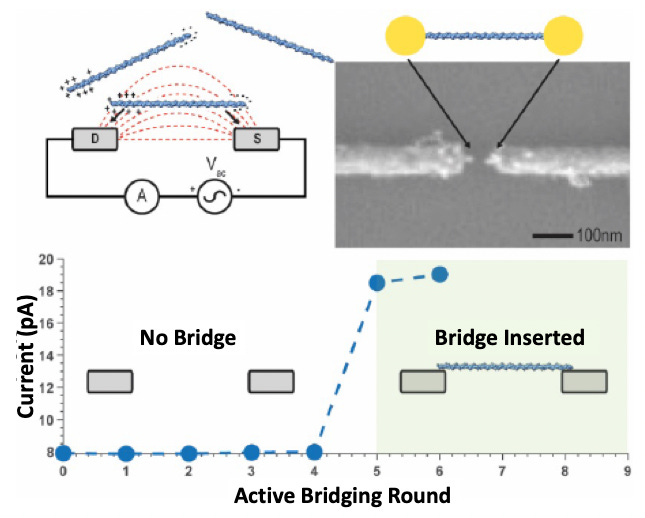
In the Roswell approach, there is indeed a nano gap, over which a “self assembling” double stranded DNA is built to form a “bridge”. With the bridge inserted, a small increase in current is detected:
Roswell propose using this bridge as a scaffold on which to build a number of molecular sensors. I’d previously assumed that the dominant source of current across the gap was due to tunneling. And it does seem that there is some small residual current, when no bridge is present. This would either need to be a tunneling current or some kind of residual ionic current.
However, after being prompted by a reader, who suggested that the dominate conduction mechanism here was conduction across the backbone, I went hunting for papers.
There does indeed seem to be academic support for this, with one publication showing currents of up to 5nA that vary across the backbone:

So it at least seems plausible that conduction across the backbone is involved. In Roswell’s approach the bridge is used as a scaffold on which to build other sensing technologies. For example they’ve shown data from a hybridization based assay:
In this case a second single stranded fragment is attached to the DNA bridge. Target complmentary strands hybridize and an increase in current is registered. I’m not clear as to why an increase would be registered if current is purely from backbone conduction.
Similarly, in their proposed sequencing approach, a polymerase is attached to the DNA bridge and again, here I’m unclear how conformational changes in the polymerase would result in a change in the backbone conductance of bridge.
The polymerase (or hybridized oligo) could be affecting the conformation of the bridge changing its ability to conduct.. though how this results in increased, rather than decreased conductance is also unclear to me.
I suspect a number of conduction paths may be present, in what is after all a relatively complex structure.
In any case, Roswell seem to have shifted away from DNA sequencing. This indicates that the approach is probably challenging.
That’s all for part 1. In part 2 I’m going to take a look at field and optical nanopore sequencing approaches. If there’s anything else I’ve missed, and you’d like me to look into, please get in touch!