Reticula Update - Thoughts On Single Molecule Colonies
After my previous adventures with Reticula (which you can read all about here) I kind of gave up on pitching it for infectious disease diagnostics.
I’m still confident that meta-genomic sequencing wins out over over qPCR in terms of its ability to detect and contain future viral outbreaks (and can be cost competitive, more sensitive).
However I’m less confident that it’s possible for me to get such a project funded in the short term.
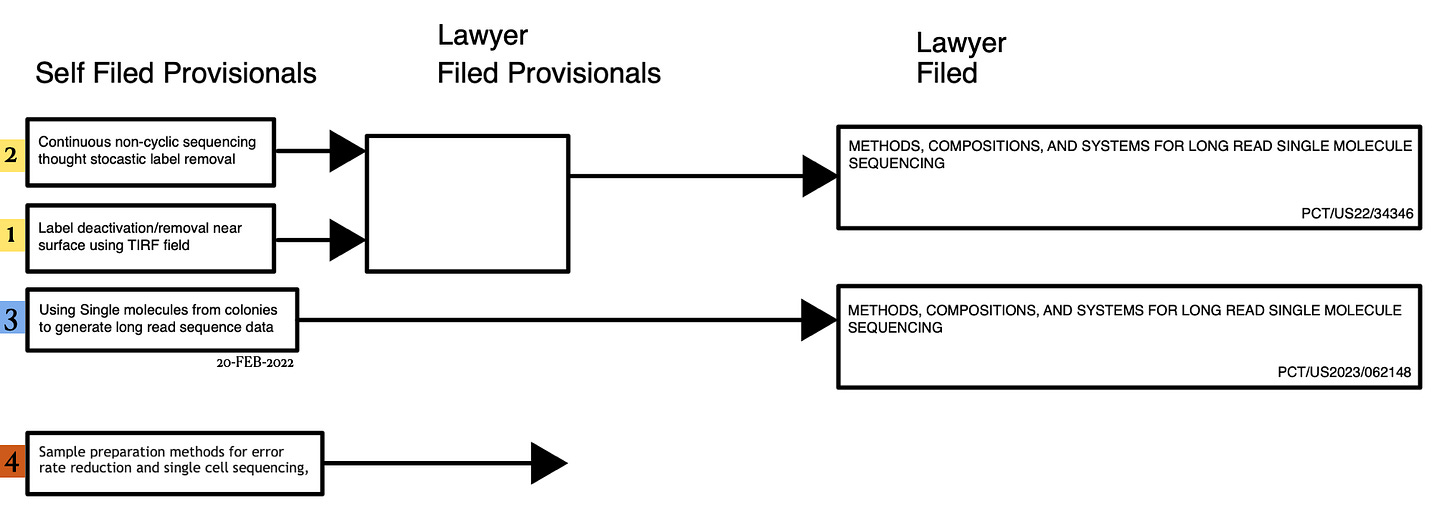
So I’ve been broadening out the Reticula. Kind of building out a portfolio of sequencing related IP:
One of the ideas I’ve been working on is a method of using cluster based sequencing to do long reads. I’ve written about this before, but now the international search report has come back:
Which seems positive!
To briefly review the approach, the idea is that we grow clusters as usual, but randomly deactivate a subset of strand in a cluster (we can terminate these strands, or potentially use other methods):
This gives us enough distance between our individual strands that they can be resolved and sequenced using single molecule approaches.
Why would we want to do this?
Well, Illumina’s current chemistry doesn’t seem to show much in terms of signal loss. This suggests that Illumina read length is currently limited by phasing (among other issues we discussed in the previous post).
Single molecule reads are not effected by phasing, they are however in general low accuracy. PacBio get round this by using CCS (cyclic consensus sequencing). In this approach we’re doing something similar to CCS but reading all the information out at once in parallel!
The approach is also compatible with both real time (PacBio) and cyclic (Illumina, Helicos) style sequencing.
But wait there’s more! While phasing is a problem in Illumina sequencing, it’s a positive advantage here. If we intentionally push reads out of phase we can readout different locations on a long template in parallel. We can therefore sub-reads covering a long fragment in parallel, which we finally assemble into high quality consensus reads.
Long reads, low error rates, fast readout.
With a few tweaks the process might even be compatible with existing sequencing instrumentation!
That’s the idea anyway. But I’m not sure what to do with it. I’ll certainly need more cash to take even the IP forward. If the idea interests you or you have further suggestions feel free to reach out (new@sgenomics.org) or on the Discord.



“ However I’m less confident that it’s possible for me to get such a project funded in the short term.”
Can you share what led you to this conclusion?